Vector Systems
Related Services
Vector CloningPlasmid DNA Preparation
Virus Packaging Services
mRNA Gene Delivery Solutions
CRISPR Genome Editing Solutions
shRNA Gene Knockdown Solutions

Regular Plasmid CRISPR Vector
CRISPR/Cas9 vectors are among several types of emerging genome editing tools that can quickly and efficiently create mutations at target sites of a genome (the other two popular ones being ZFN and TALEN).
Cas9 is a member of a class of RNA-guided DNA nucleases which are part of a natural prokaryotic immune system that confers resistance to foreign genetic elements such as plasmids and bacteriophage. Within the cell, the Cas9 enzyme forms a complex with a guide RNA (gRNA), which provides targeting specificity through direct interaction with homologous 18-22nt target sequences in the genome. Hybridization of the gRNA to the target site localizes Cas9, which then cuts the target site in the genome.
To achieve CRISPR-mediated gene targeting it is essential for the target cells to co-express Cas9 and the target site-specific gRNA at the same time. This can be accomplished by either expressing both Cas9 and the gRNA sequence from the same vector (a.k.a. all-in-one vector) or by using separate vectors for driving Cas9 and gRNA expression (Cas9 only and gRNA only vectors, respectively). The advantage of using an all-in-one vector for expressing Cas9 and gRNA is that it provides the opportunity to deliver all the required components for CRISPR-mediated gene editing to the cell using a single vector which is technically straight forward. Using separate vectors for expressing Cas9 and gRNA requires co-transfection of the target cells with two separate vectors which can be technically challenging since not all cells will be transfected with both gRNA and Cas9 vectors simultaneously. An alternative approach for using separate vectors is to transfect cells or organisms stably expressing high-level of Cas9 with the desired gRNA sequences. However, this method can be considerably time-consuming and labor intensive. Our all-in-one regular plasmid CRISPR vector helps to circumvent the mentioned challenges by expressing Cas9 and the desired gRNA sequence from a single regular plasmid vector.
Our regular plasmid CRISPR vector is a highly efficient tool for conventional transfection-based delivery of both Cas9 and the target site-specific gRNA sequence into a variety of mammalian cells. Delivering plasmid vectors into mammalian cells by conventional transfection is one of the most widely used procedures in biomedical research. While a number of more sophisticated gene delivery vector systems have been developed over the years such as lentiviral vectors, adenovirus vectors, AAV vectors and piggyBac, conventional plasmid transfection remains the workhorse of gene delivery in many labs. This is largely due to its technical simplicity as well as good efficiency in a wide range of cell types. A key feature of transfection with regular plasmid vectors is that it is transient, with only a very low fraction of cells stably integrating the plasmid in the genome (typically less than 1%).
The regular plasmid CRISPR vector is available for expressing either single-gRNA or dual-gRNAs. While the single-gRNA vector is widely used for conventional CRISPR genome editing applications such as generating single gene knockouts, dual-gRNA vectors are necessary for applications requiring simultaneous targeting of a pair of genomic sites. Examples of such applications include: 1) paired Cas9 nickase experiments where the “nickase” mutant form (hCas9-D10A) of hCas9 is used in conjunction with two gRNAs targeting the two opposite strands of a single target site to generate single strand cuts one on each strand, thereby leading to a DSB with increased targeting specificity than a single gRNA; 2) generating deletion of a fragment between two DSBs targeted by a pair of gRNAs; and 3) targeting two different genes simultaneously. While the single gRNA vector consists of a single human U6 promoter driving the target site-specific gRNA sequence, the dual gRNA vector consists of two consecutive U6 promoters driving the expression of gRNA sequences specific to two genomic target sites of interest.
Two variants of Cas9 enzyme are available in our all-in-one regular plasmid CRISPR vectors. The standard humanized Cas9 (hCas9) variant efficiently generates double-strand breaks (DSBs) at target sites, while the “nickase” mutant form (hCas9-D10A) generates only single-stranded cuts in DNA. If hCas9-D10A nickase is used in conjunction with two gRNAs targeting the two opposite strands of a single target site, then the nickase enzyme will generate single strand cuts on both strands, resulting in DSBs at the target site (as described above). This approach generally reduces off-target effects of CRISPR/Cas9 expression because targeting by both gRNAs is necessary for DSBs to be generated.
Cellular repair of DSBs by the nonhomologous end-joining pathway (NHEJ) usually results in small deletions, or more rarely insertions and base substitutions. When these mutations disrupt a protein-coding region (e.g. a deletion causing a frameshift), the result is a functional gene knockout. Alternatively, and less efficiently, DSBs can be repaired by homology-directed repair (HDR), using exogenous donor DNA template, which is co-introduced with the CRISPR/Cas9 vector. This can result in replacement of the target genomic DNA sequence with template sequence, generating small targeted base changes, such as point mutations. Nicked genomic DNA also frequently undergoes homology-directed repair (HDR), and if exogenous template DNA is introduced into the cell along with a targeted hCas9-D10A nickase, then small base changes can be generated.
Most DNA sequence can be effectively targeted using the CRISPR/Cas9 system. However, there is a strict requirement for an NGG (sometimes NAG) sequence, known as protospacer adjacent motif (PAM), which is located on the immediate 3’ end of the gRNA recognition sequence within the target DNA.
Click to view user testimonials about our CRISPR vectors
For further information about this vector system, please refer to the papers below.
| References | Topic |
|---|---|
| Science 339:819-23 (2013) | Description of genome editing using the CRISPR/Cas9 system |
| Cell. 154:1380–9 (2013) | Use of Cas9 D10A double nicking for increased specificity |
| Nat. Biotech. 31:827–832 (2013) | Specificity of RNA-guided Cas9 nucleases |
Highlights
Our regular plasmid CRISPR vector is optimized for high copy number replication in E. coli and high-efficiency transfection. Cells transfected with the vector can be selected and/or visualized based on marker gene expression as chosen by the user. The regular plasmid CRISPR vector system is designed to deliver Cas9 and a target site-specific gRNA sequence using a single regular plasmid vector. This vector is available for expressing either single-gRNA or dual-gRNAs enabling users to target either one or two genomic target sites of interest depending upon their experimental goal.
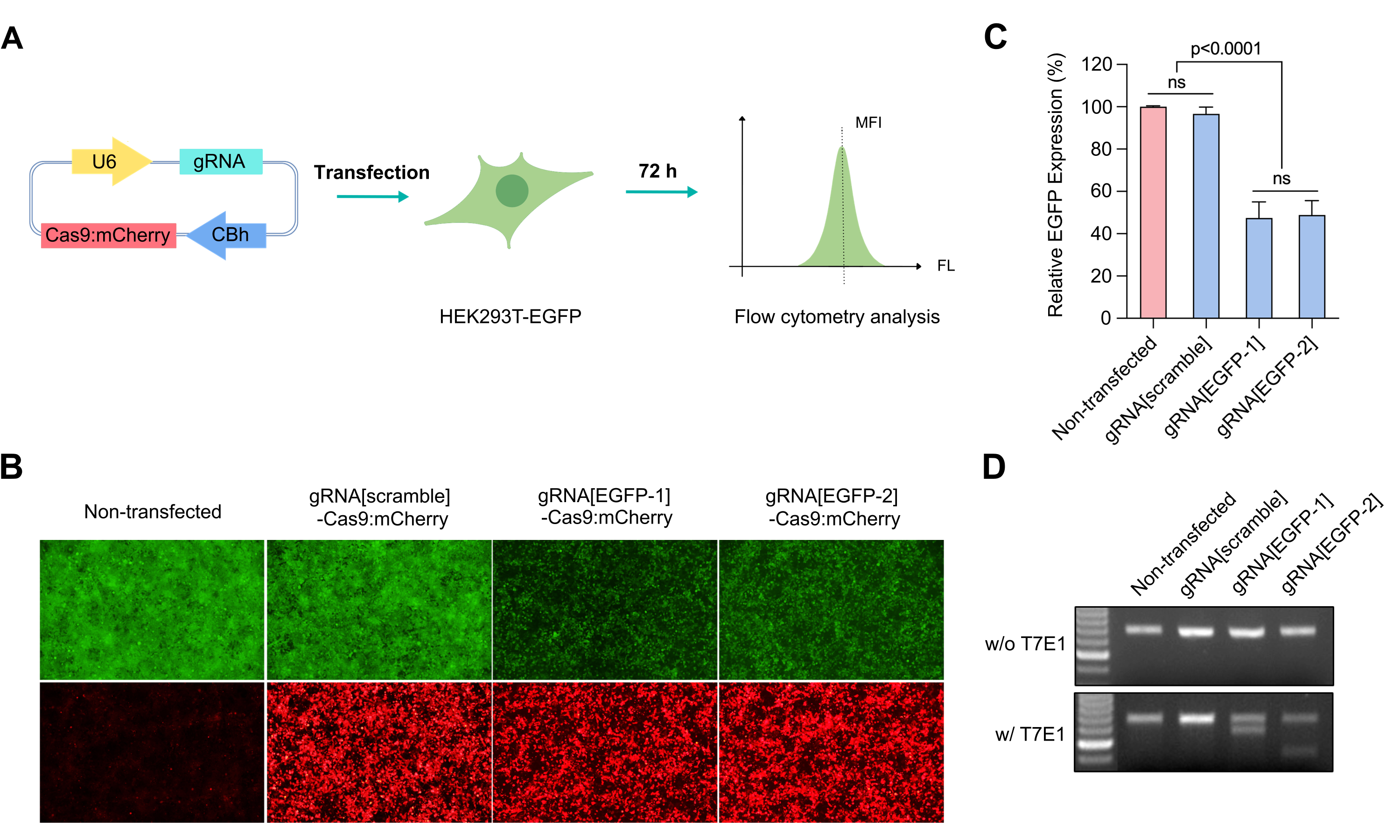
Figure 1. Gene editing with all-in-one CRISPR system. (A) Regular plasmid expressing Cas9:T2A:mCherry and EGFP-targeting or scramble gRNA was transfected into EGFP stable expressing HEK293T cells (HEK293T-EGFP). The expression of EGFP was observed and measured by flow cytometry at 72 h post transfection. MFI stands for mean fluorescence intensity. (B) EGFP and mCherry expression was observed by microscopy (100X). (C) Relative EGFP expression in all-in-one CRISPR vector transduced cells compared to the non-transfected control. Relative EGFP expression was calculated by [MFI (experimental group) – MFI (WT HEK293T cells)] / [MFI (HEK293T-EGFP cells) – MFI (WT HEK293T cells)]. Mean±SD, ns P>0.05, ANOVA with Tukey’s post hoc test. (D) The gRNA-targeted region was PCR amplified from the genomic DNA, and the editing was confirmed using the T7E1 assay.
Advantages
Simplicity: The simple homology relationship between the gRNA and the target makes the CRISPR/Cas9 system conceptually simple and easy to design. Our regular plasmid CRISPR vector system is designed for delivering both Cas9 as well as the target site-specific gRNA sequence to mammalian cells. This provides the opportunity to deliver all the required components for CRISPR-mediated gene editing to the target cells using a single regular plasmid vector which is technically straight forward and less time-consuming than using two separate vectors for Cas9 and gRNA delivery.
Transient expression: Transfection of the CRISPR/Cas9 plasmid vector results in strong transient expression of the Cas9 protein and gRNA within the target cells. Without drug selection, the plasmid will be lost over time eliminating the Cas9 and gRNA from the target cells after genome editing has taken place.
Disadvantages
Lower specificity: Some off-target activity has been reported for the CRISPR/Cas9 system, and in general the TALEN system has lower off-target activity than CRISPR/Cas9. However, off-target effects can be significantly mitigated by using the mutant version hCas9-D10A nickase in conjunction with two gRNAs to target the two opposite strands of a single target site to generate single strand cuts one on each strand, thereby leading to a DSB with increased targeting specificity than a single gRNA used in conjunction with the wild type hCas9 nuclease.
PAM requirement: CRISPR/Cas9 based targeting is dependent on a strict requirement for a protospacer adjacent motif (PAM), located on the immediate 3’ end of the gRNA recognition sequence.
U6 Promoter: This drives high level expression of the gRNA. This is the promoter of the human U6 snRNA gene, an RNA polymerase III promoter which efficiently expresses short RNAs.
gRNA: Guide RNA compatible with Cas9 derived from Streptococcus pyogenes.
Terminator: Terminates transcription of the gRNA.
CBh promoter: Chicken beta-actin promoter. Drives expression of the downstream Cas9 nuclease.
Cas protein: Cas9 nuclease variant chosen by user.
BGH pA: Bovine growth hormone polyadenylation signal. It facilitates transcriptional termination of the upstream ORF.
CMV promoter: Human cytomegalovirus immediate early promoter. It drives the ubiquitous expression of the downstream marker gene.
SV40 late pA: Simian virus 40 late polyadenylation signal. It facilitates transcriptional termination of the upstream ORF.
Ampicillin: Ampicillin resistance gene. It allows the plasmid to be maintained by ampicillin selection in E. coli.
pUC ori: pUC origin of replication. Plasmids carrying this origin exist in high copy numbers in E. coli.
U6 Promoter: This drives high level expression of the gRNA. This is the promoter of the human U6 snRNA gene, an RNA polymerase III promoter which efficiently expresses short RNAs.
gRNA #1: The first guide RNA compatible with Cas9 derived from Streptococcus pyogenes.
gRNA #2: The second guide RNA compatible with Cas9 derived from Streptococcus pyogenes.
Terminator: Terminates transcription of the gRNA.
CBh promoter: Chicken beta-actin promoter. Drives expression of the downstream Cas9 nuclease.
Cas protein: Cas9 nuclease variant chosen by user.
BGH pA: Bovine growth hormone polyadenylation signal. It facilitates transcriptional termination of the upstream ORF.
CMV promoter: Human cytomegalovirus immediate early promoter. It drives the ubiquitous expression of the downstream marker gene.
SV40 late pA: Simian virus 40 late polyadenylation signal. It facilitates transcriptional termination of the upstream ORF.
Ampicillin: Ampicillin resistance gene. It allows the plasmid to be maintained by ampicillin selection in E. coli.
pUC ori: pUC origin of replication. Plasmids carrying this origin exist in high copy numbers in E. coli.
