mRNA Gene Delivery Solutions
VectorBuilder provides a one-stop solution for the development of mRNA-based therapeutics, such as vaccines, gene editing, chimeric antigen receptor (CAR), and protein expression in cells or embryos. Based on extensive design and production experience, our team can support researchers for in vitro transcription (IVT) vector design, vector cloning, in vitro mRNA synthesis, LNP-mRNA production, and in vitro/in vivo functional testing to accelerate the development of mRNA-based vaccines and gene therapy. For large-scale manufacturing of mRNA therapeutics, check out our CDMO services. For purchasing ready-to-use mRNA products, check out our catalog IVT mRNA and LNP-mRNA.
Service Details
 IVT vector design & cloning
IVT vector design & cloning
- Royalty-free IVT backbone with no IP constraints for commercial use
- Sequence optimization for achieving high expression
- 5’ and 3’ UTR
- Coding sequence
- Kozak
- Robust cloning and transcription of 120 nt (and longer) polyA tail for high expression
 IVT mRNA & LNP manufacturing
IVT mRNA & LNP manufacturing
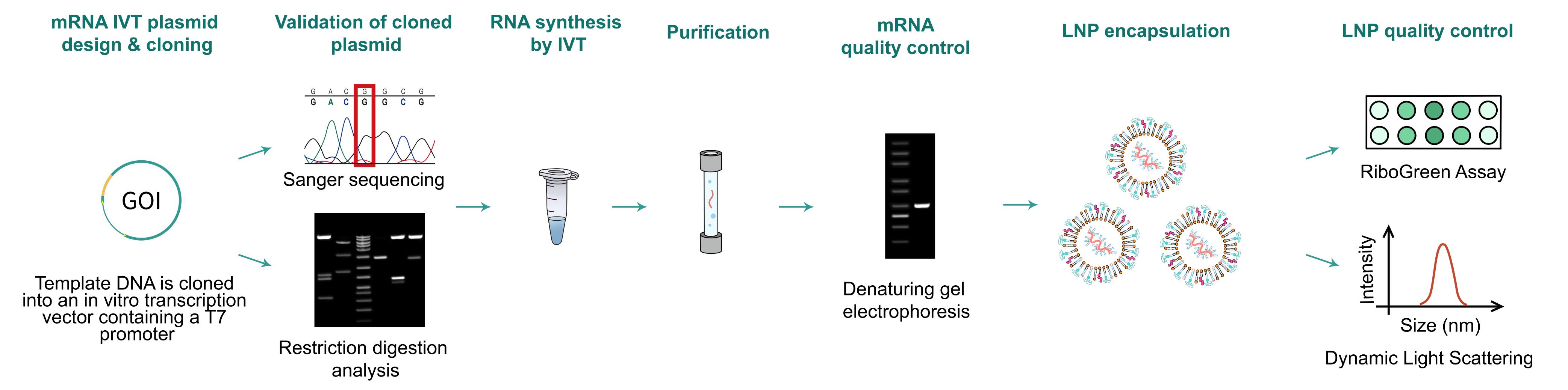
Figure 1. Typical workflow of mRNA synthesis and LNP encapsulation.
- From vector cloning to LNP encapsulation in as fast as 5 weeks
- Synthesis of mRNA and self-amplifying RNA (saRNA) of up to 10,000 nt from ug to gram scale
- High capping efficiency (up to 99%) by co-transcriptional or enzymatic capping
- Modified nucleotides for reducing immune response and achieving robust expression in vivo
- m1Ψ
- m5C
- 5moU
- Proprietary purification technology to rapidly and efficiently remove impurities include:
- IVT DNA template
- Partially transcribed mRNA products
- Residual protein
- Double-stranded RNA (dsRNA)
- High-quality LNP with conventional and custom formulation:
- High encapsulation efficiency
- Low polydispersity index (PDI)
- Improved stability
- Increased delivery efficiency
- Antibody-conjugation compatibility
 Quality control (QC)
Quality control (QC)
VectorBuilder offers an extensive variety of QC methods for IVT mRNA and LNP encapsulation. Default QC items (marked with √) are always performed while optional QC items are performed depending on individual project needs.
IVT mRNA
LNP encapsulation
| Attribute | QC Assay | Research-grade | GMP-like | |
|---|---|---|---|---|
| Identity | mRNA sequence | Sanger sequencing | √ | √ |
| mRNA length | Denaturing agarose gel electrophoresis | √ | √ | |
| Capillary gel electrophoresis (CGE) | Optional | √ | ||
| General/physical property | mRNA concentration | UV spectrophotometry | √ | √ |
| RiboGreen assay | Optional | √ | ||
| Appearance | Visual inspection | Optional | √ | |
| Potency | Gene expression | In vitro translation followed by Western blot | Optional | Optional |
| Cell transfection | Optional | Optional | ||
| Safety | Sterility | Bioburden test | Optional | √ |
| Mycoplasma | Culture method | Optional | √ | |
| qPCR | Optional | Optional | ||
| Endotoxin | Kinetic chromogenic assay (KCA) | Optional | √ | |
| Purity | mRNA integrity | Denaturing agarose gel electrophoresis | √ | √ |
| Capillary gel electrophoresis (CGE) | Optional | √ | ||
| A260/280 | UV spectrophotometry | √ | √ | |
| Capping efficiency | LC-MS | Optional | √ | |
| PolyA analysis | LC-MS | Optional | √ | |
| Residual dsRNA | Dot blot assay | Optional | √ | |
| Residual plasmid DNA | qPCR | Optional | √ | |
| Residual protein | NanoOrange assay | Optional | √ | |
| Residual solvents | Gas chromatography | Optional | Optional | |
| Attribute | QC Assay | Research-grade | GMP-like |
|---|---|---|---|
| Encapsulation efficiency | RiboGreen assay | √ | √ |
| Particle size, PDI | Dynamic light scattering (Zetasizer) | √ | √ |
| Surface charge | Dynamic light scattering (Zetasizer) | √ | √ |
 Functional validation
Functional validation
- Assess the effects of different sequence optimizations (UTRs, coding sequence, polyA, Kozak, etc.) on GOI expression in parallel, using our high-throughput cloning, synthesis, and testing platforms.
- Established functional validation platforms for various applications, such as antigen presentation, antibody expression, CAR expression, and CRISPR.
- Assess LNP-mRNA gene delivery efficacy and safety using animal models including rodents and non-human primates (NHPs).
Technical Information
IVT vector sequence optimization
- UTR
- Coding sequence
- PolyA tail
- Kozak sequence
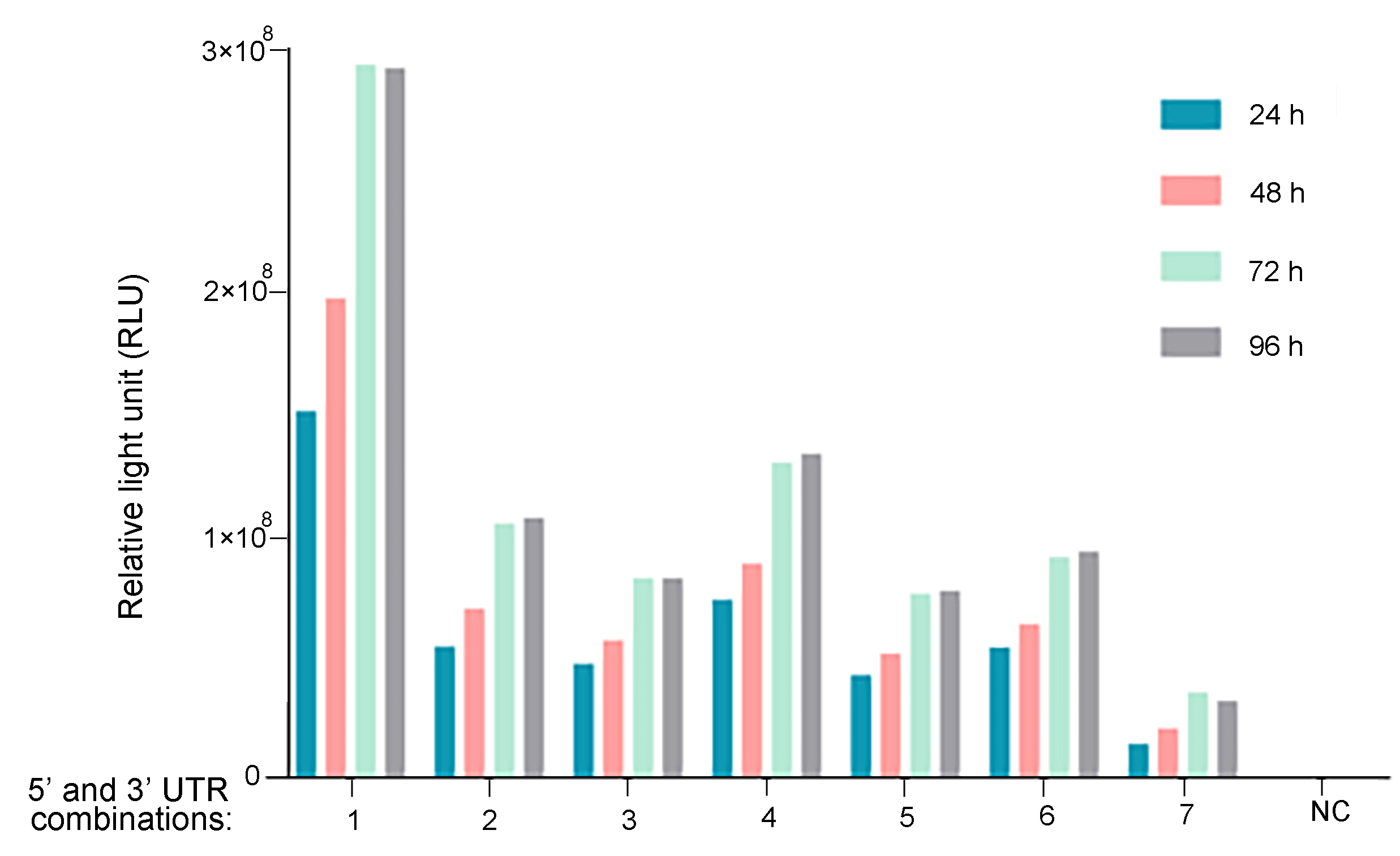
Figure 2. UTR optimization for improved mRNA expression. Different 5' and 3' UTR combinations were tested for regulating Guassia luciferase expression in vitro. 293T cells were seeded on 12-well plates at a density of 2.3x105 cells per well. Cells were transfected with 1 ug of mRNA per well. At 6 h, 24 h, 48 h, 72 h, and 96 h post-transfection, Gaussia luciferase activities were measured from cell culture medium.
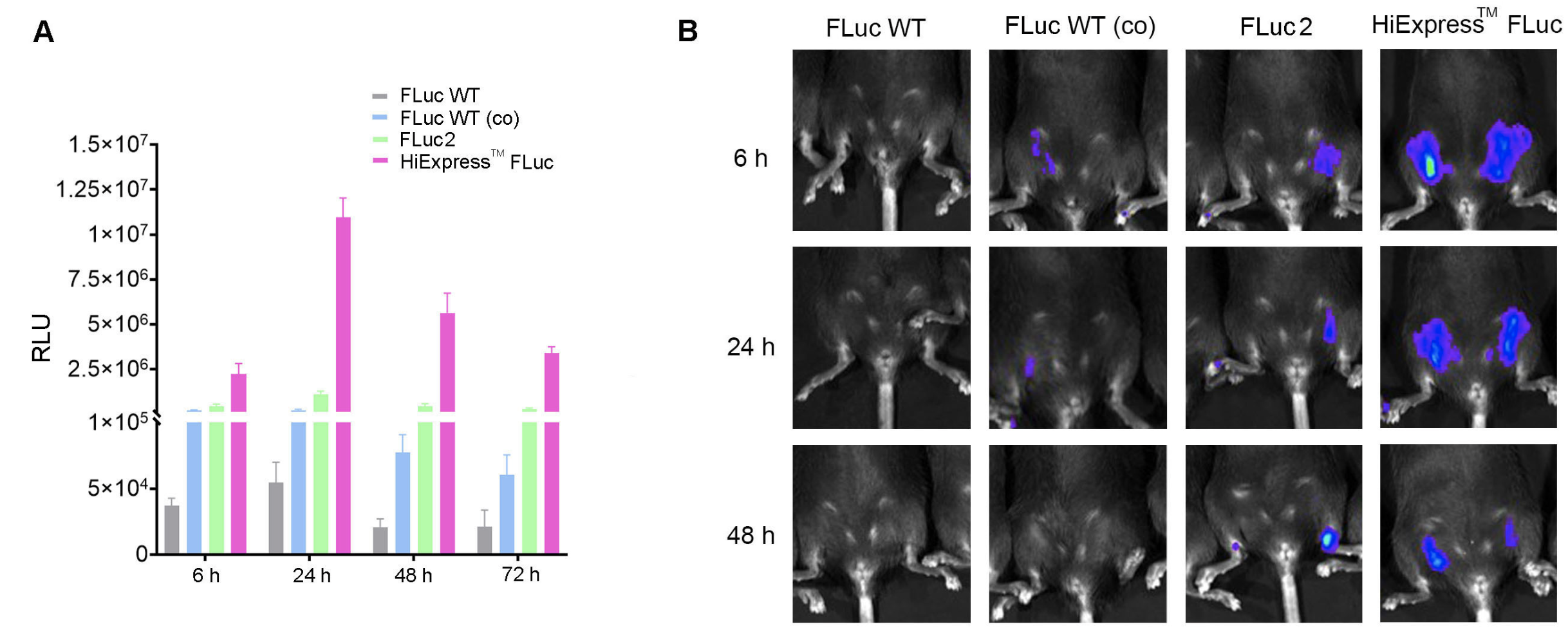
Figure 3. Codon optimization increases mRNA expression in vitro and in vivo. (A) Expression of HiExpress™ Firefly Luciferase mRNA and other luciferase mRNA in HEK293T cells. Cells grown on a 12-well plate were transfected with 0.5 ug of mRNA per well and luciferase activity was measured at 6 h, 24 h, 48 h, and 72 h post-transfection. (B) Luciferase activity measured in adult C57BL/6 mice injected intramuscularly with 30 ug of LNP encapsulated mRNA at 6 h, 24 h, and 48 h post-injection. FLuc WT indicates wild-type firefly luciferase. FLuc WT (co) indicates codon-optimized wild-type firefly luciferase. FLuc2 indicates Luc2 firefly luciferase.
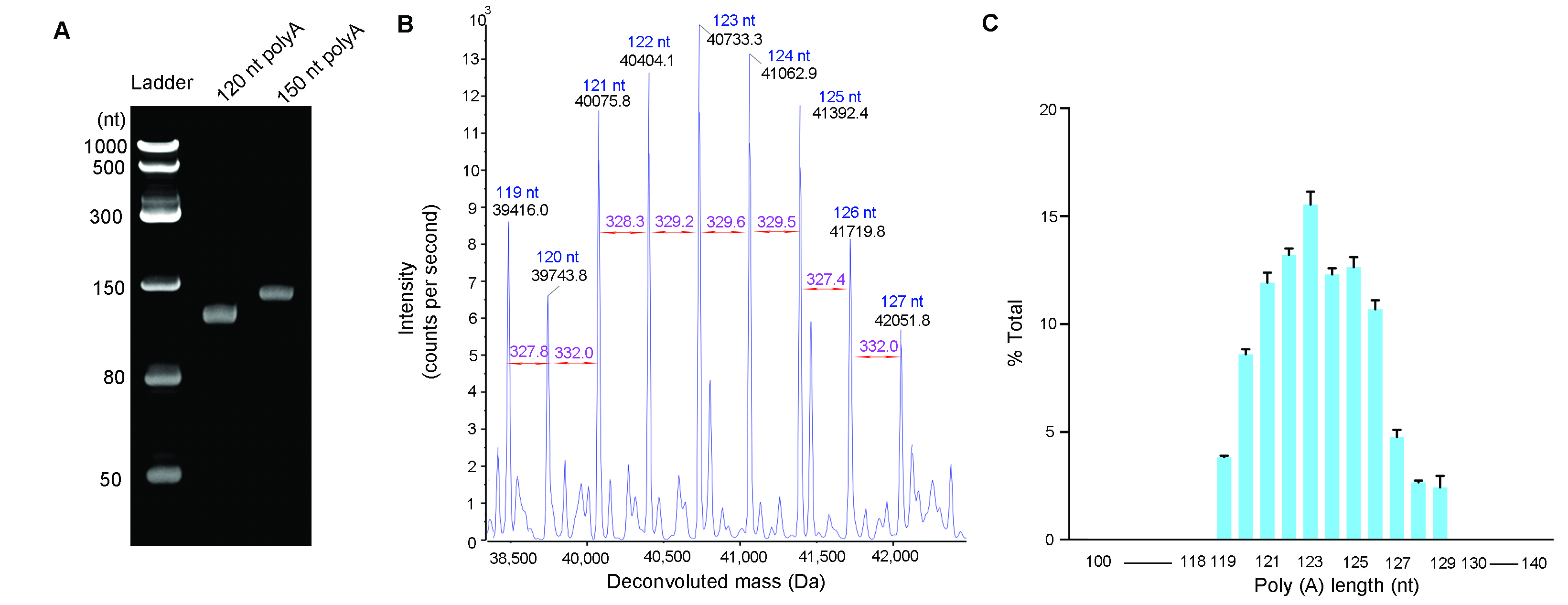
Figure 4. PolyA tail size analysis. PolyA tails were cleaved from IVT mRNA using ribonuclease T1 and isolated by oligo dT affinity chromatography. (A) Isolated polyA tails analyzed by Urea-PAGE gel electrophoresis. 60 ng of polyA tails with expected size of 120 nt and 150 nt were loaded on denaturing Urea-PAGE gel, respectively. (B) Isolated polyA tails analyzed by LC-MS. Deconvoluted spectrum was generated from a 120 pmol of polyA tails with an expected size of 120 nt. (C) Size distribution of the polyA tails with an expected size of 120 nt. The bar graph demonstrates the polyA tails have small heterogeneity. The error bars represent standard deviation from technical triplicates. The weighted average length is 123 nt.
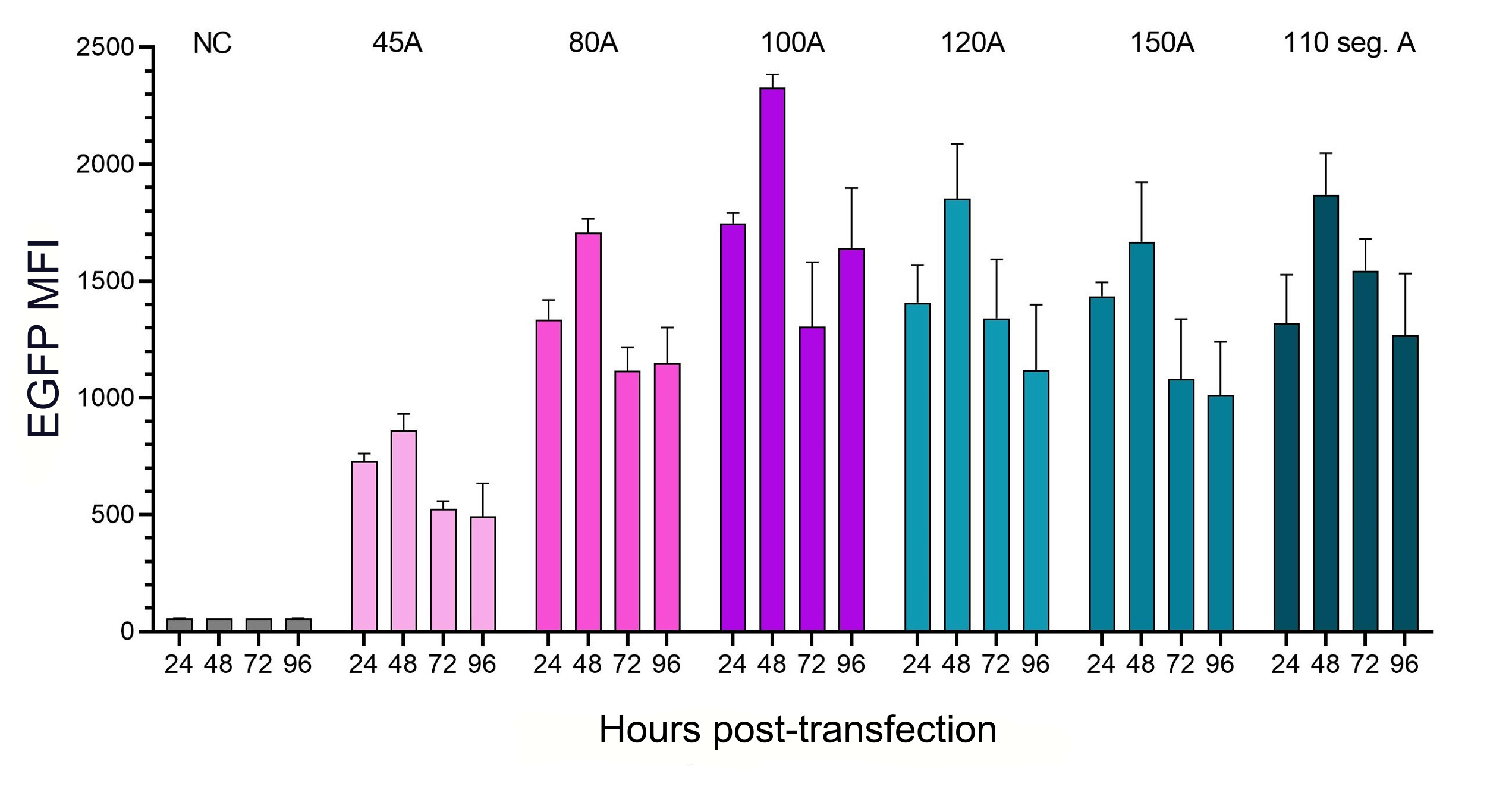
Figure 5. The influence of polyA length and structure on translational efficiency. HEK293T cells were transfected with IVT EGFP mRNA with the same Cap1 and UTRs but different polyA tail lengths as listed above.
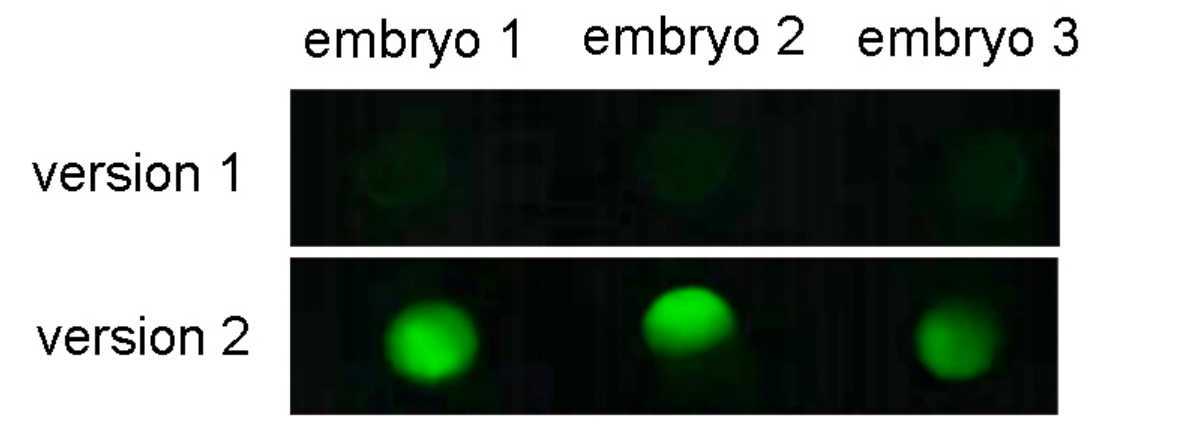
Figure 6. Kozak sequence optimization increased IVT EGFP mRNA expression in zebrafish. One-cell stage zebrafish embryos were measured for EGFP expression at 6 h post-microinjection of 250 pg of zebrafish EGFP IVT mRNA. Fluorescent images indicated optimized Kozak sequence (version 2) greatly improved EGFP expression in vivo.
IVT mRNA synthesis optimization
- IVT mRNA integrity
- Co-transcriptional and enzymatic capping
- Nucleotide modification
- dsRNA removal
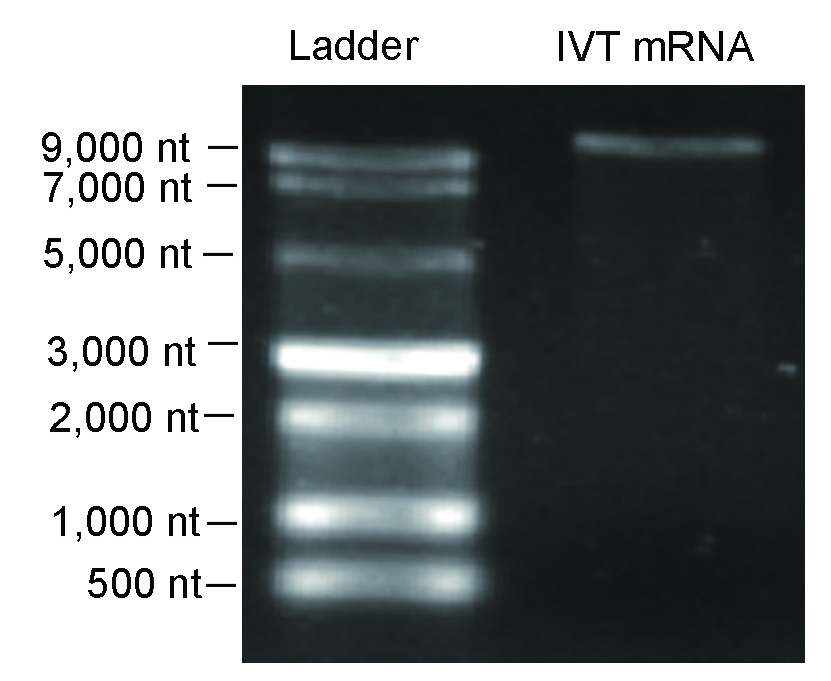
Figure 7. Denaturing agarose gel result indicated high integrity was achieved for >10,000 nt IVT mRNA.
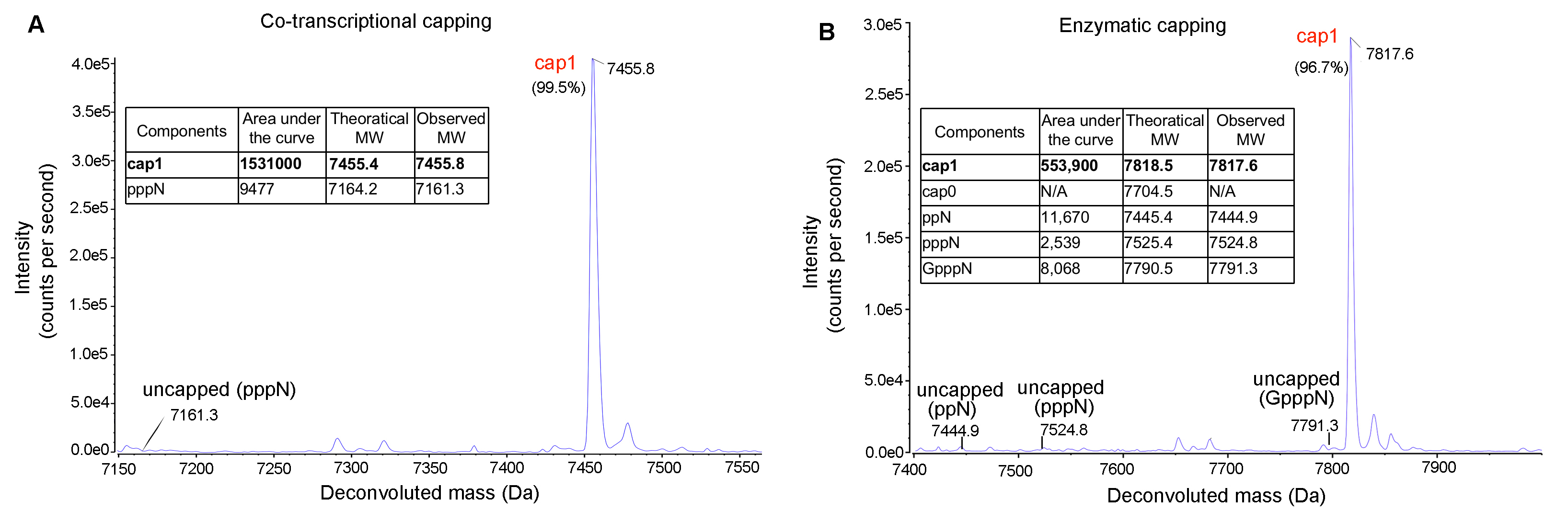
Figure 8. IVT mRNA capping efficiency analyzed by LC-MS. Highly efficient capping (>99%) can be achieved either using (A) co-transcriptional or (B) enzymatic approaches.
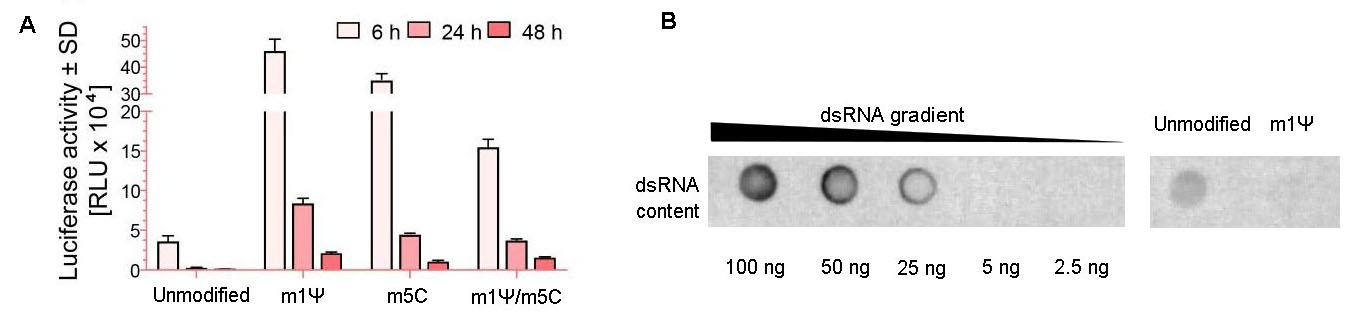
Figure 9. Modified nucleotides increased mRNA expression and decreased dsRNA impurities. (A) Expression of firefly luciferase in 293T cells. The mRNA was generated with or without modified nucleotides, N1-Methylpseudouridine (m1Ψ) and 5-Methylcytosine (m5C). Cells grown on the 12-well plates were transfected with 1 ug of mRNA per well. Luciferase activities in 293T cells at 6 h, 24 h, and 48 h post-transfection were measured. Error bars indicate standard deviations. (B) Equal amount (750 ng per dot) of magnetic bead-purified EGFP IVT mRNA with or without nucleotide modification (m1Ψ) was blotted and subsequently detected by dot blot assay for estimating the dsRNA impurity.
As a major trigger of undesired immunogenicity, double-stranded RNA (dsRNA) is a by-product of IVT. Dot blot result demonstrates extra purification steps (e.g. IP-PR) may be necessary for achieving an ultra-purification scale with very low levels of dsRNA.
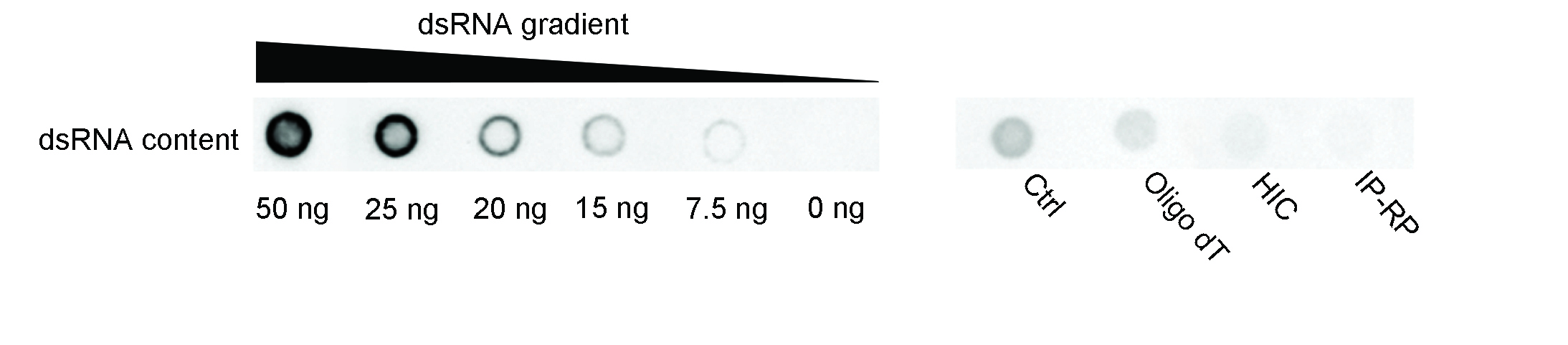
Figure 10. dsRNA removal efficiency of different purification processes. Equal amounts (1500 ng per dot) of hSpCas9 IVT mRNA purified by different processes were blotted and subsequently detected by dot blot assay for estimating the dsRNA impurity. Abbreviations: HIC, Hydrophobic interaction chromatography; IP-RP, Ion-pair reversed-phase liquid chromatography.
LNP-mRNA QC data
VectorBuilder has optimized our encapsulation technology to ensure the homogeneity, stability and efficacy of our LNP encapsulated mRNA.
- TEM
- PDI and zeta potential
Cryo-transmission electron microscopy (cryo-TEM) results show good structural integrity and size homogeneity of our LNP-mRNA.
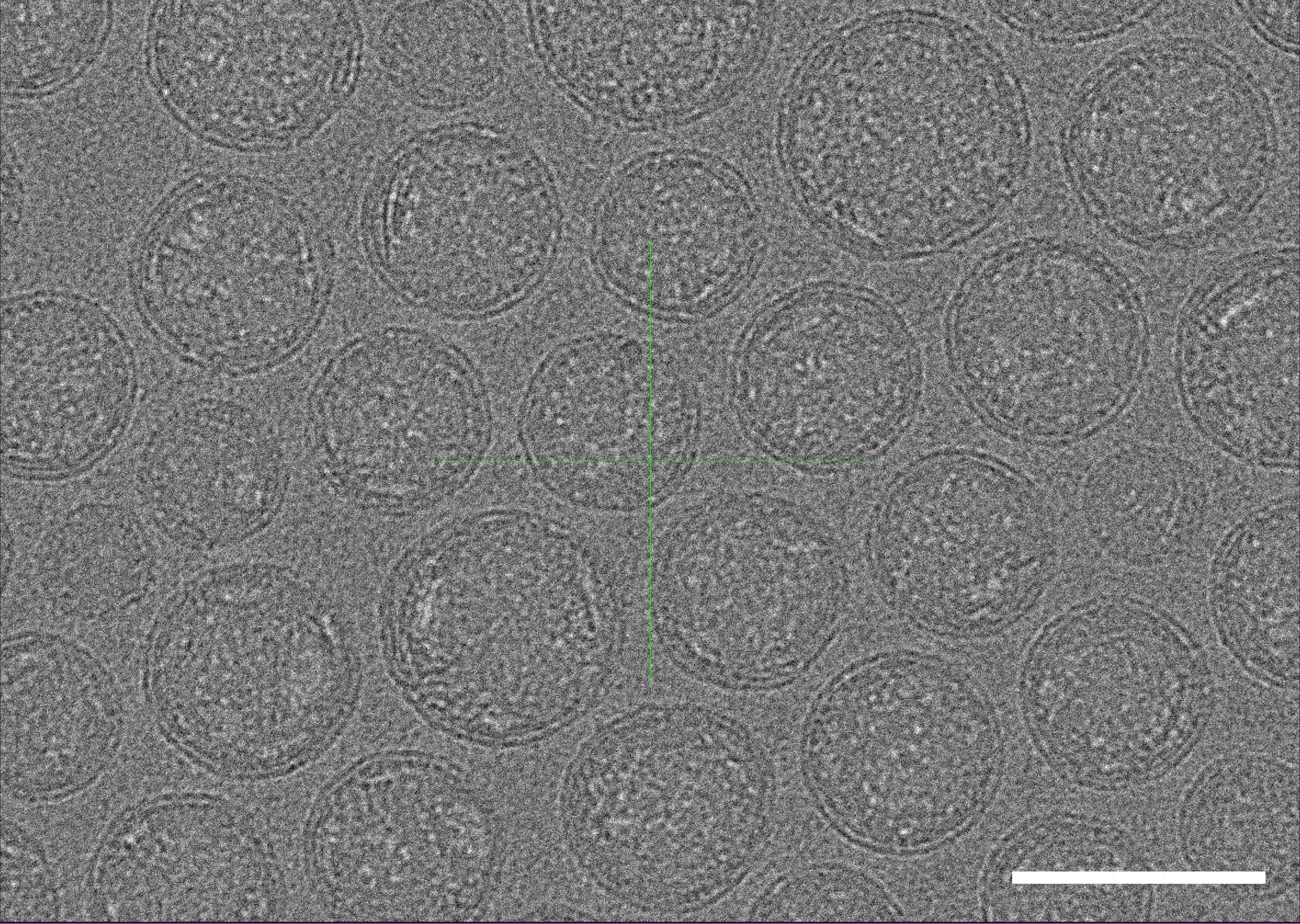
Figure 11. Cryo-TEM micrographs of LNP-mRNA. Scale bar=100 nm.
Dynamic light scattering (DLS) analysis shows our LNP-mRNA products can reach a very low PDI (as low as PDI<0.1).
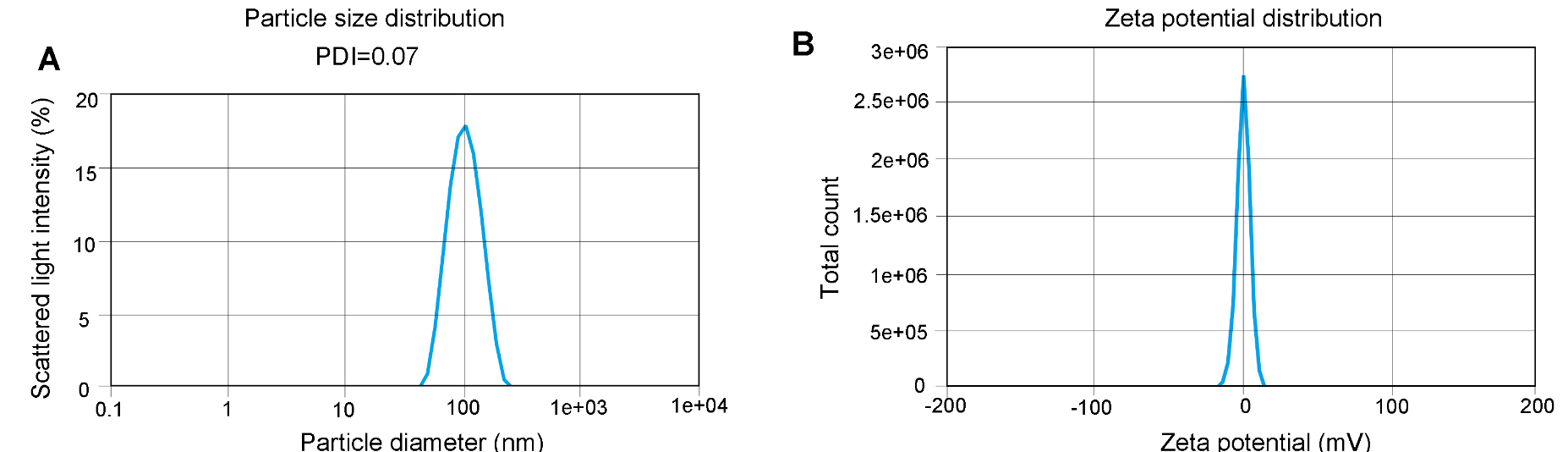
Figure 12. Particle size and Zeta potential distribution analysis. PDI (A) and Zeta potential (B) was determined by DLS which measures the intensity differences of fluctuated light due to motion of particles. The Zeta potential of the sample is between -1.872 mV and +1.872 mV.
LNP-mRNA functional validation
- LNP-mRNA expression in vitro
- LNP-mRNA expression in vivo
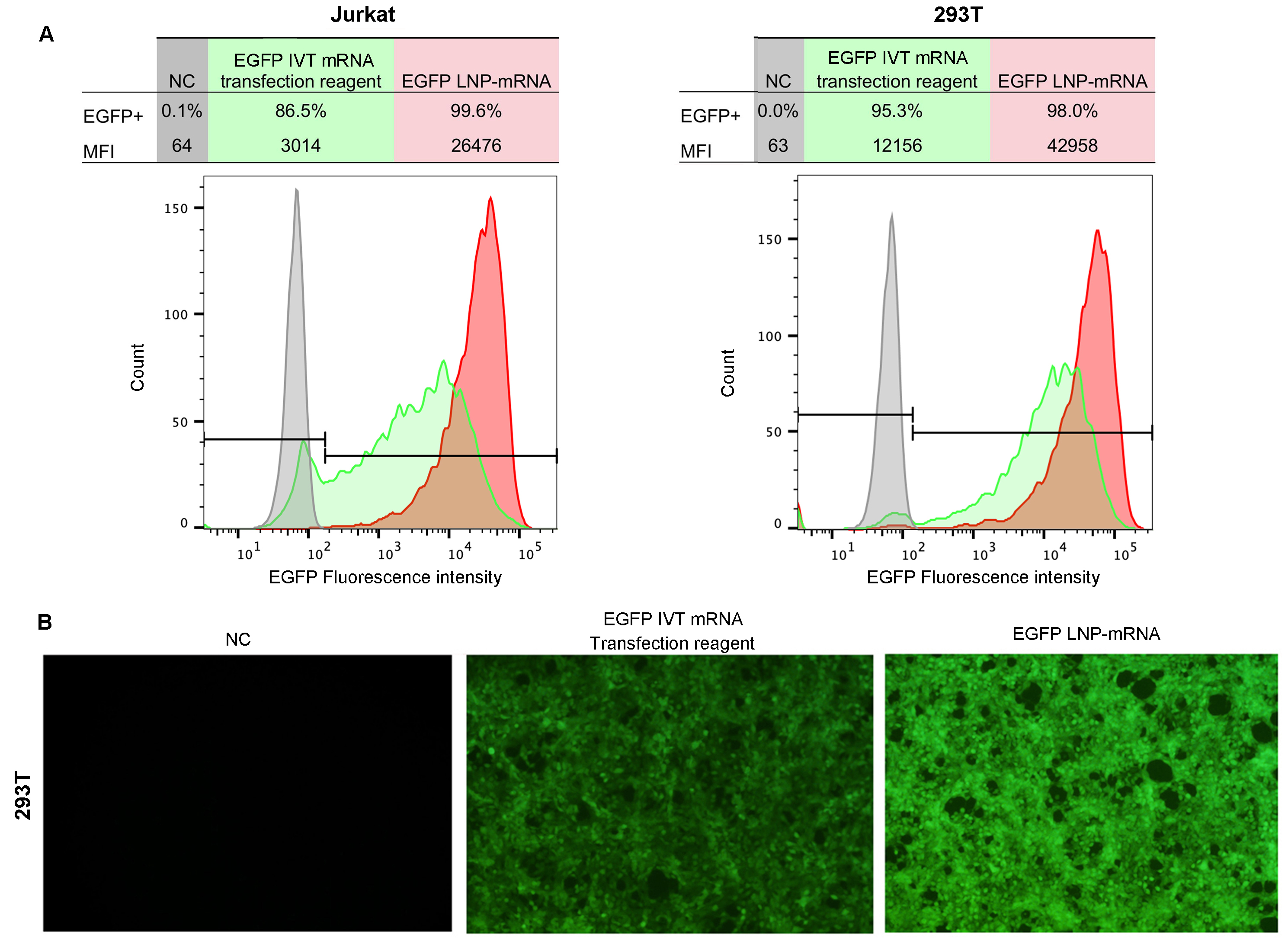
Figure 13. Efficient mRNA delivery and expression using LNP in vitro. Cells were transfected with LNP encapsulated EGFP mRNA or EGFP mRNA mixed with commercial transfection reagent. (A) Flow cytometry analysis of EGFP expression in Jurkat and 293T cells and (B) Fluorescent imaging of 293T cells at 24 hours post-transfection. MFI: median fluorescence intensity.
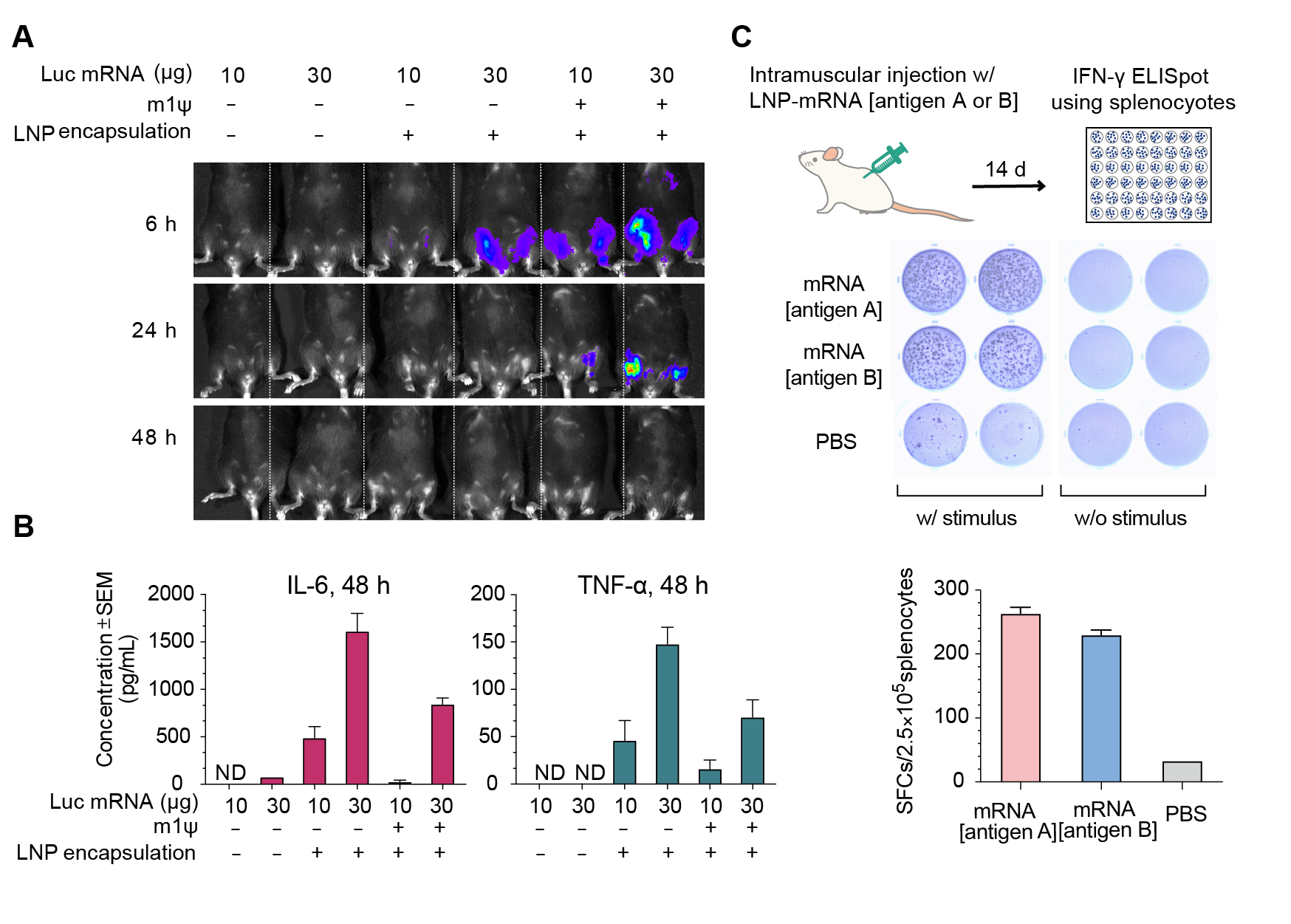
Figure 14. Expression of luciferase (Luc) mRNA and mRNA induced immune response in mice. (A) Luciferase activity visualized by live imaging at 6 h, 24 h, and 48 h post-injection. (B) Two pro-inflammatory cytokines, IL-6 and TNF-⍺, were quantified in the serum at 48 h post-injection. Error bars represent standard errors. Mice strain: C57BL/6J; mice age: 8 weeks; injection method: intramuscular injection. (C) IFN-γ ELISpot assay of splenocytes derived from Balb/C mice 14 days post intramuscular injection of 30 ug LNP-encapsulated mRNA coding for viral antigen A, viral antigen B, or control PBS.
Antibody-conjugated LNP-mRNA
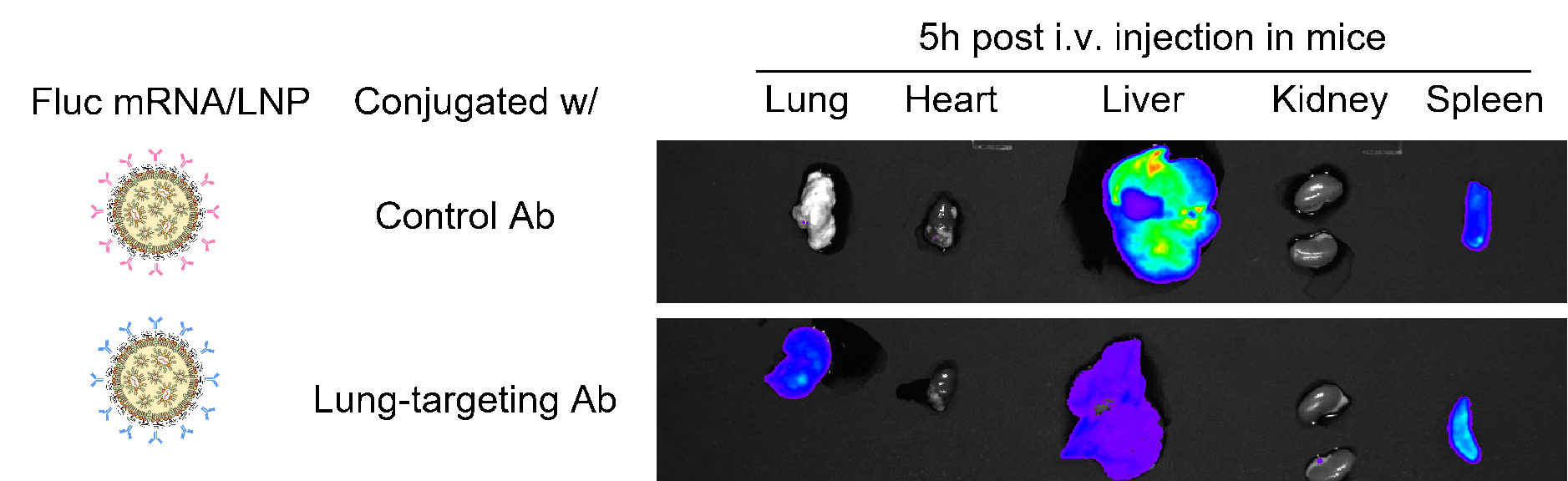
Figure 15. Anti-CD31 conjugated firefly luciferase (FLuc) LNP-mRNA showed improved luciferase expression in lung. Mice strain: C57BL/6J; mice age: 6-8 weeks; mice gender: female; administration route: tail vein. Negative controls: IgG2a-conjugated FLuc LNP-mRNA and naked FLuc mRNA.
Documents
Material Safety Data Sheet (MSDS) Certificate of Analysis (COA) User Instructions Brochures & FlyersHow to Order
FAQ
Cap 0 refers to N7-methylguanosine (m7G) that is added to the 5’ end of eukaryotic mRNAs via a 5’ to 5’ triphosphate linkage. This modification is added via a series of enzymatic reactions that occur co-transcriptionally and functions to regulate nuclear export, transcript stability, and promotes translation of the mRNA through recognition by eukaryotic translation initiation factor (eIF4E). Cap 1 refers to the addition of a methyl group to the 2’O on the first nucleotide (m7GpppNm) of the transcribed mRNA sequence in addition to the m7G cap. In mammalian cells, cap 1 structure is an important marker for mRNA to be recognized as self and not targeted by innate immunity. Adding cap 1 structure to synthesized mRNA has been demonstrated to enhance mRNA expression in vivo and reduced its immunogenicity.
Capping for in vitro transcribed RNA can occur either co-transcriptionally with cap analogs or post-transcriptionally via enzymatic reactions. We offer both capping methods, and the efficiency of them has been well validated using LC-MS. Depending on the client-preferred capping method, we will choose a compatible backbone for cloning the IVT mRNA vector from the beginning.
Cells contain cytosolic and endosomal RNA receptors that activate the immune response upon recognition of foreign RNA. Modified nucleotides are commonly found in endogenous cellular RNA. Incorporating certain modified nucleotides in IVT mRNA reduces its immunogenicity, alters secondary structure, and increases translation efficiency and half-life in a sequence-dependent manner. We provide a wide range of modified nucleotides, including the commonly used N1-Methylpseudouridine (m1Ψ) and 5-Methylcytosine (m5C). N1-Methylpsuedouridine and 5-Methylcytosine are naturally occurring nucleotides that were first identified in tRNAs, however, their use in coding mRNAs has only recently been appreciated. These methylated derivatives of uridine and cytosine can replace their canonical nucleotides in mRNA IVT and translation without altering traditional Watson-Crick base pairing. A major advantage to their use in mRNA therapeutics is their ability to alter recognition by RNA immune receptors thus mitigating unwanted immune effects and enhancing transcript stability and translation.









