Mutation Library Construction
VectorBuilder offers extensive expertise in constructing custom mutagenesis libraries. These libraries are critical for many studies as the initial step for probing the key residues and domains in proteins. We provide versatile strategies for creating your diverse pool of mutations, including chip-based synthesis, error-prone PCR, degenerate codon utilization, and DNA shuffling. These mutations can be confined to a single region or distributed across multiple regions along the sequence.
Highlights
- Diverse cloning strategies: Tailored to your research goals, we employ various approaches to construct your mutation library.
- High complexity: We can generate mutation libraries exceeding 108 in complexity.
- Great coverage and high uniformity: Our libraries typically achieve >98% coverage of targeted variants.
Service Details
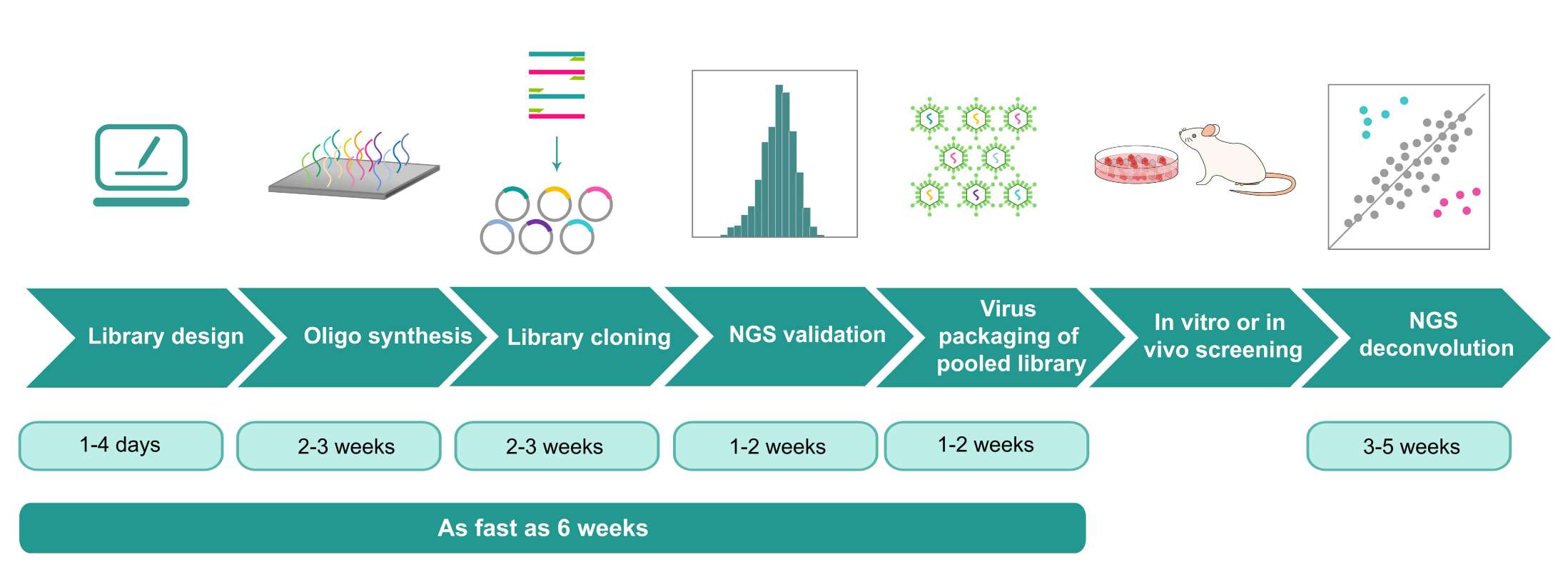
Price and turnaround Price Match
| Service Module | Brief Description | Price (USD) | Turnaround |
|---|---|---|---|
| Mutation library design | VectorBuilder's team of highly experienced scientists will design your customized mutation library using versatile strategies. You have the option to select from our extensive collection of validated vector backbones. | Free | 1-4 days |
| Pooled library cloning | Includes oligo/gene synthesis, massive parallel cloning of inserts for variable regions into desired vector backbone, preliminary validation by Sanger sequencing, and full validation by NGS. Deliverable includes library in E. coli glycerol stock and NGS report. | From $2,300 | 5-8 weeks |
| Virus packaging of pooled library | Please click here to view detailed info of virus packaging services. The price for packaging pooled library is 1.5-fold of the price for packaging single vector. | ||
| NGS deconvolution of post-screening sample | Includes NGS library preparation from genomic DNA of screened cells, Illumina sequencing (>500x coverage), and data analysis. | From $320 per sample | 3-5 weeks |
Technical Information
Mutation library construction strategies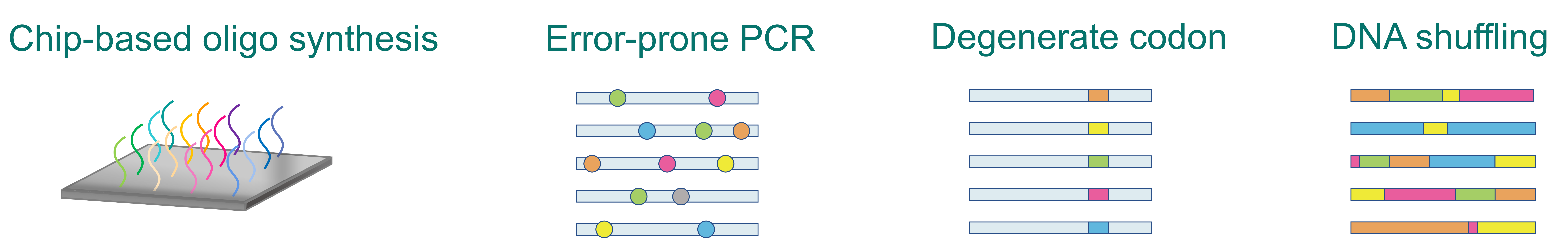
Chip-based oligo synthesis
Chip-based oligo synthesis is ideal for generating pre-designed sequence variants of various sizes and sequences. This includes saturation mutations at any given site or deep mutational scanning libraries for entire proteins or specific protein fragments. Our chip-based DNA synthesis can typically reach up to 300 nucleotides with a low error rate.
Error-prone PCR
Error-prone PCR is the most straightforward approach for creating highly variable mutation libraries which involves modifying standard PCR methods to mutagenize the original sequence. More specifically, error-prone PCR employs a combination of various sub-optimal PCR conditions including low fidelity polymerases, longer extension times, higher Mg2+ concentrations, the addition of Mn2+, and varying dNTP concentrations for introducing random point mutations into the original sequence.
Degenerate codon
The degeneracy of genetic code allows one amino acid to be encoded by multiple codons. Degenerate codons in oligonucleotide synthesis introduce controlled, highly randomized mutations. This approach enables targeted focus on regions and amino acids that are likely to yield positive hits. The NNK library, for example, a widely used degenerate mutation library, involves NNK degenerate primers encompassing 32 codon combinations (N= A/C/G/T, K= G/T), which encode all 20 amino acids and one stop codon. Utilizing degenerate codons in library construction is an effective way to introduce diversity.
| Degenerate Codon | # Codons | Stop Codon | Amino Acids |
|---|---|---|---|
| NNN | 64 | 3 | All 20 |
| NNK / NNS | 32 | 1 | All 20 |
| NDT | 12 | 0 | RNDCGHILFSYV |
| DBK | 18 | 0 | ARCGILMFSTWV |
| NRT | 8 | 0 | RNDCGHSY |
DNA shuffling
DNA shuffling is a highly efficient method to generate chimeric DNA sequences through homologous recombination. This technique involves the digestion of parental gene(s) into random fragments, followed by their reassembly into new variants using primer-less PCR, which relies on partial sequence homology for recombination. Alternatively, synthetic shuffling can be employed for creating high-complexity libraries, combining rational design with directed evolution. DNA family shuffling may also be employed following other mutagenesis methods such as error-prone PCR.
Experimental data
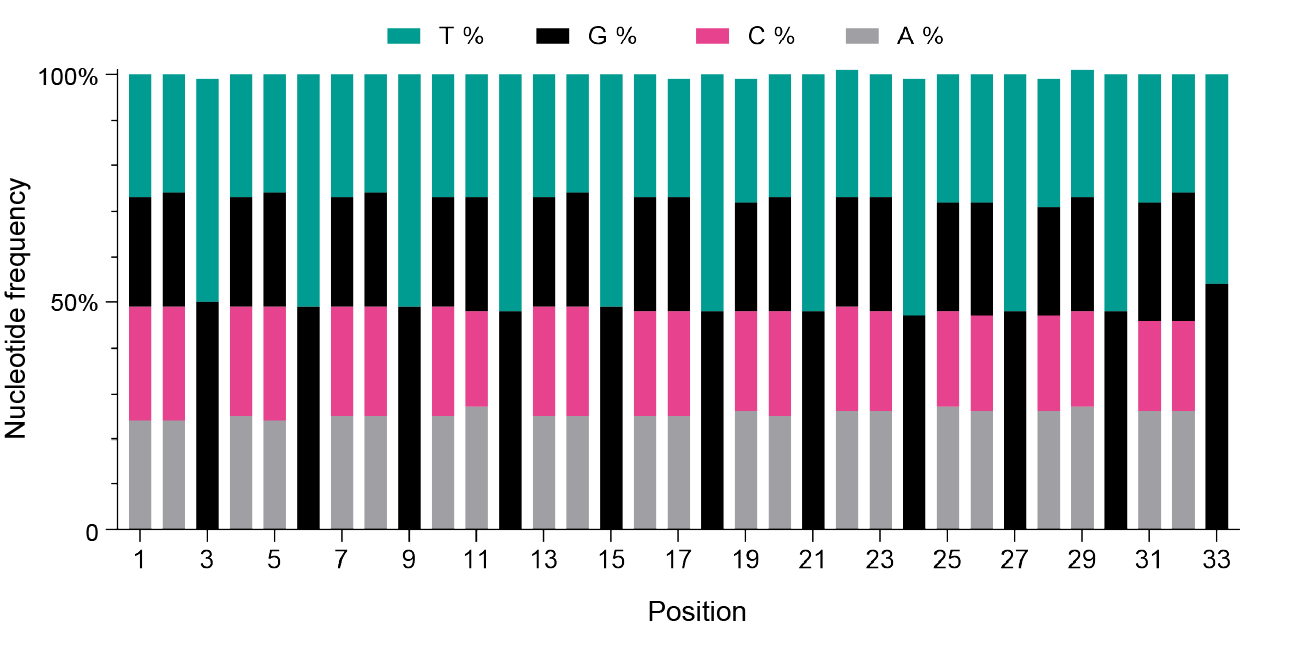
Figure 1. Nucleotide composition of an (NNK)11 plasmid library. The library was constructed using the NNK degenerated codon strategy. Each bar represents a distinct nucleotide position along the DNA sequence, with colors indicating the observed nucleotides. All positions exhibited anticipated nucleotides with observed frequencies close to theoretical values (N = 25% A + 25% T + 25% G + 25% C, K = 50% T + 50% G).
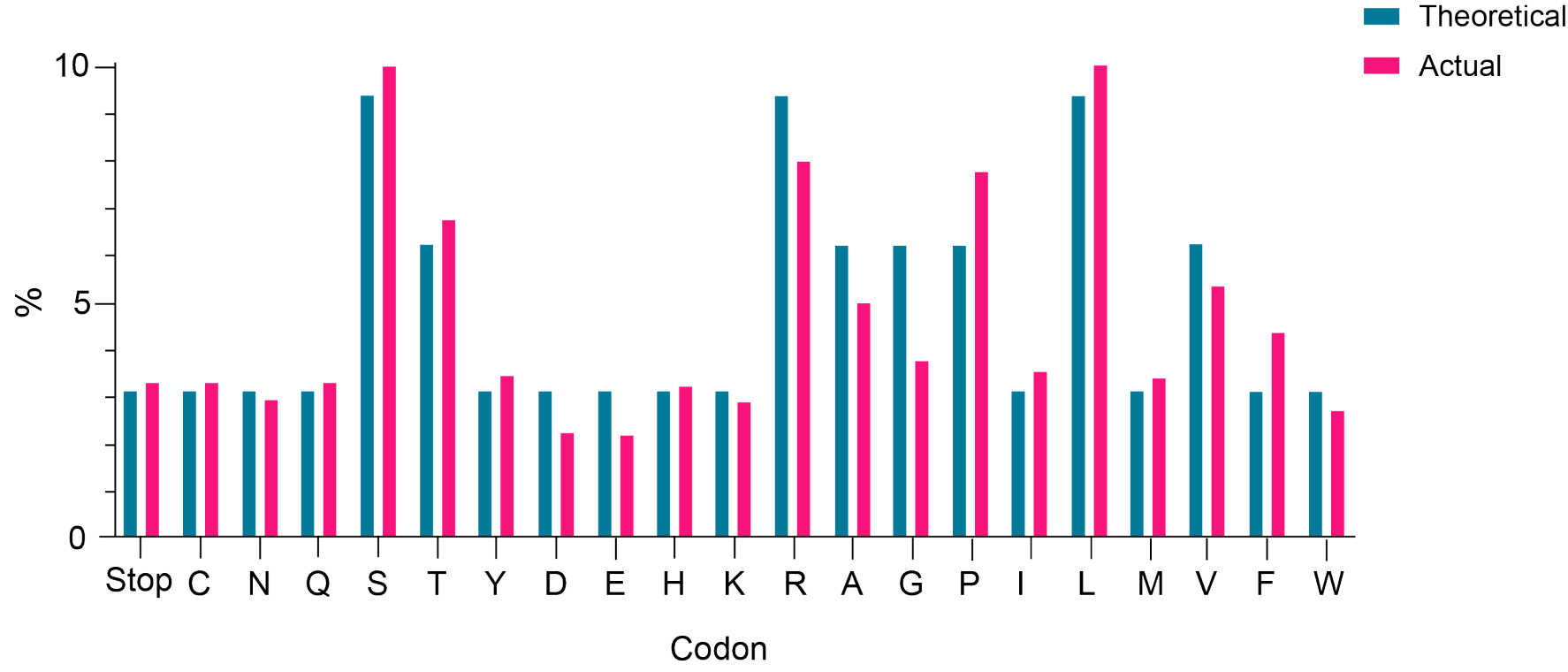
Figure 2. Distribution of amino acids in a 7-mer variable region of a plasmid library. The library was constructed using the NNK degenerate codon strategy. Each bar represents the theoretical (teal) or actual (pink) percentage of specific amino acids or the stop codon. The observed frequencies of all 20 amino acids and the stop codon closely matched the theoretical frequencies.
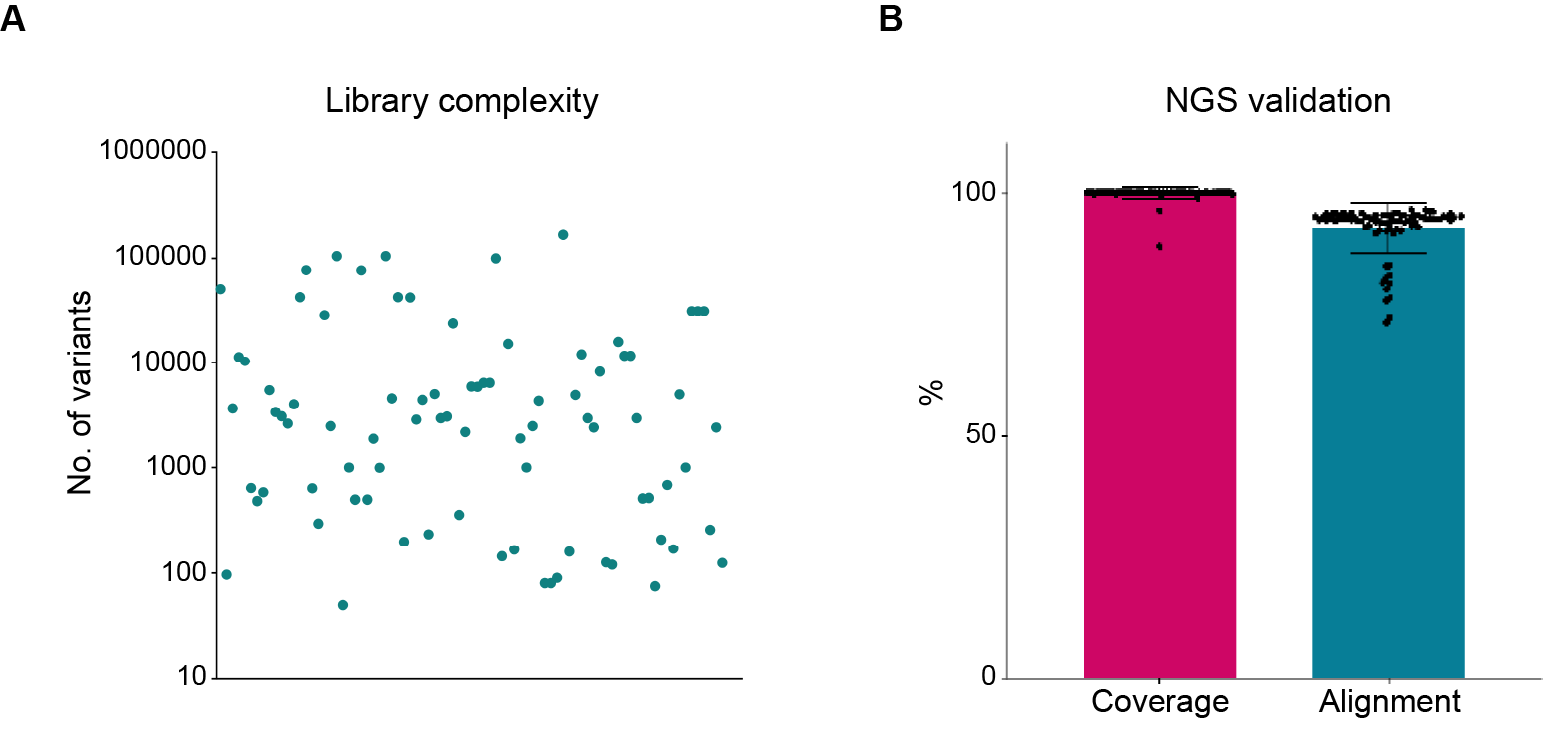
Figure 3. Complexity, coverage, and alignment rate of chip-based oligo libraries constructed by VectorBuilder. (A) The library complexity ranged from 102 to exceeding 105. (B) Both the coverage and read alignment rate were very high, highlighting the exceptional coverage and low error rate of our libraries.
How to Order
Customer-supplied library plasmid pool
If the customer-supplied premade plasmid pools are used, please send us the materials following the Materials Submission Guidelines. Please strictly follow our guidelines to set up shipment to avoid any delay or damage of materials. All customer-supplied materials undergo mandatory QC by VectorBuilder which may incur a $100 surcharge for each item. Please note that production may not be initiated until customer-supplied materials pass QC. For customer-supplied premade plasmid pools, we cannot provide any guarantees regarding the complexity or uniformity of the library.
Resources
FAQ
| Strategy | Advantages | Limitations |
|---|---|---|
| Error-prone PCR |
|
|
| Degenerate codon |
|
|
| DNA shuffling |
|
|



