Enhancer/Promoter Screening
Enhancers and promoters are non-coding genomic regions that play crucial roles in regulating the spatial and temporal patterns of gene expression. Identifying these cis-regulatory elements, analyzing their gene regulation activities, and engineering novel regulatory variants are critical for understanding and optimizing gene expression, with profound implications in gene therapy. VectorBuilder offers comprehensive solutions for researchers and drug developers to effectively identify functional enhancers/promoters through massive in-parallel screening, both in vitro and in vivo.
Request design support now to get a free consultation.
Highlights
- Streamlined full-service platform: Comprehensive solutions including library and screening strategy design, library construction, virus packaging, in vitro and in vivo screening, NGS analysis, hit identification, and functional characterization.
- Expertise in library design: High signal-to-noise ratio, barcode incorporation, NGS readout strategy and more, for powerful, unbiased, and versatile screening.
- High-complexity libraries with high-titer, high-purity virus packaging: Libraries (>108) available in non-viral, lentiviral, AAV, piggyBac and other delivery systems. Virus packaging includes lentivirus (>108 TU/ml) and AAV (>1013 GC/ml).
- Multiple in vivo models including non-human primate (NHP): Available models for screening include mice, rats, and two non-human primate species: crab-eating macaque (Macaca fascicularis, a.k.a. cynomolgus monkey) and rhesus macaque (Macaca mulatta). All animal experiments are conducted in AAALAC-accredited facilities.
Service Workflow
Below is a typical workflow of enhancer/promoter screening:
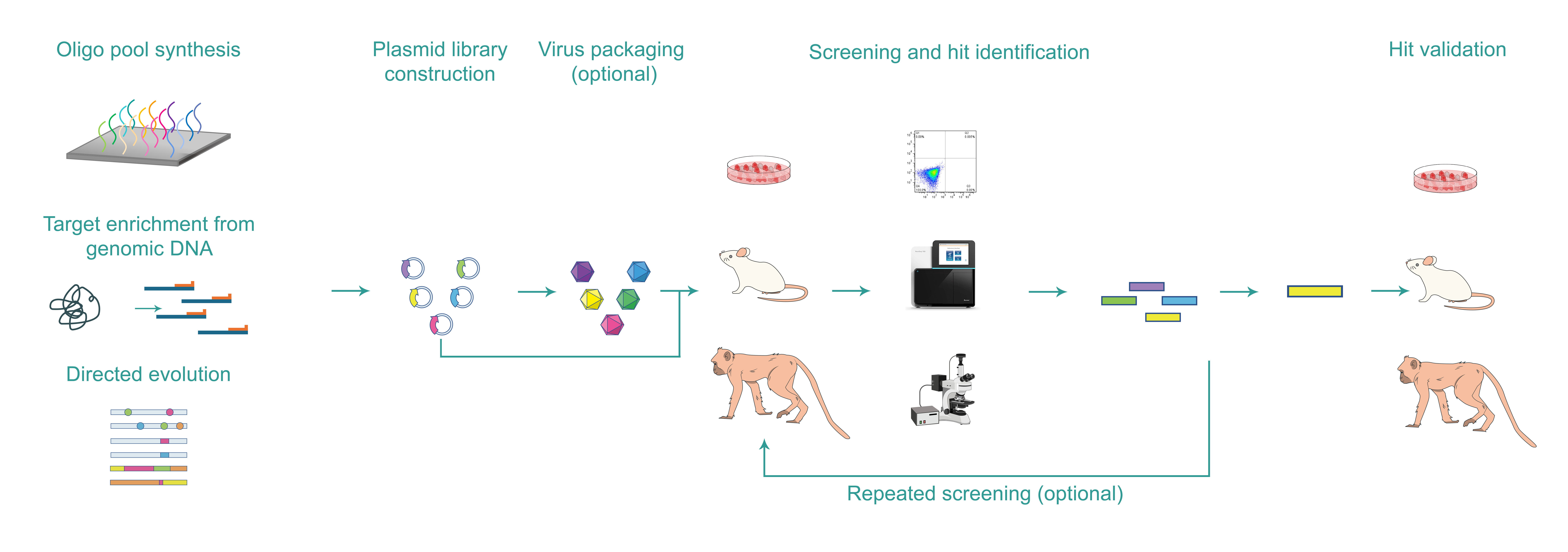
Enhancer/promoter library design and construction
In vitro/in vivo screening
Hit validation
Library design
The figures below illustrate typical vector designs for enhancer (Figure 1) and promoter (Figure 2) libraries.

Figure 1. Key vector components for an enhancer library.
Enhancer: The enhancer sequence to be screened can be inserted either upstream or downstream of the reporter gene. When the enhancer is positioned downstream (A) of the reporter gene and before the poly(A) signal, it is transcribed alongside the reporter gene and, therefore, can be measured directly by mRNA-seq. When positioned upstream (B) of the reporter gene, a barcode sequence is commonly incorporated in the 3’ UTR region of the reporter gene and can be transcribed with the reporter gene. The barcode serves as the proxy of the enhancer and can be sequenced by mRNA-seq.
Minimal promoter: A user-selected minimal promoter sequence is placed here. This will drive transcription of the downstream reporter if an enhancer element is present to activate the transcription. In the absence of such enhancer, the minimal promoter will be almost completely inactive.
Click to view more information about commonly used minimal promoters
Kozak: Kozak consensus sequence. It is placed in front of the start codon of the ORF of interest because it is believed to facilitate translation initiation in eukaryotes.
Reporter: A visually detectable bright fluorescent protein gene (such as GFP) or a chemiluminescent protein gene (such as luciferase). This allows highly sensitive detection of enhancer activity.
Barcode: Enhancer library barcodes are unique, short sequences within individual plasmids. After screening, the read count of each barcode is determined through NGS analysis. By correlating the unique barcode sequences with the specific enhancer variants, the transcriptional activity of particular enhancers can be identified.
SV40 late pA: Simian virus 40 late polyadenylation signal. It facilitates transcriptional termination of the upstream ORF.
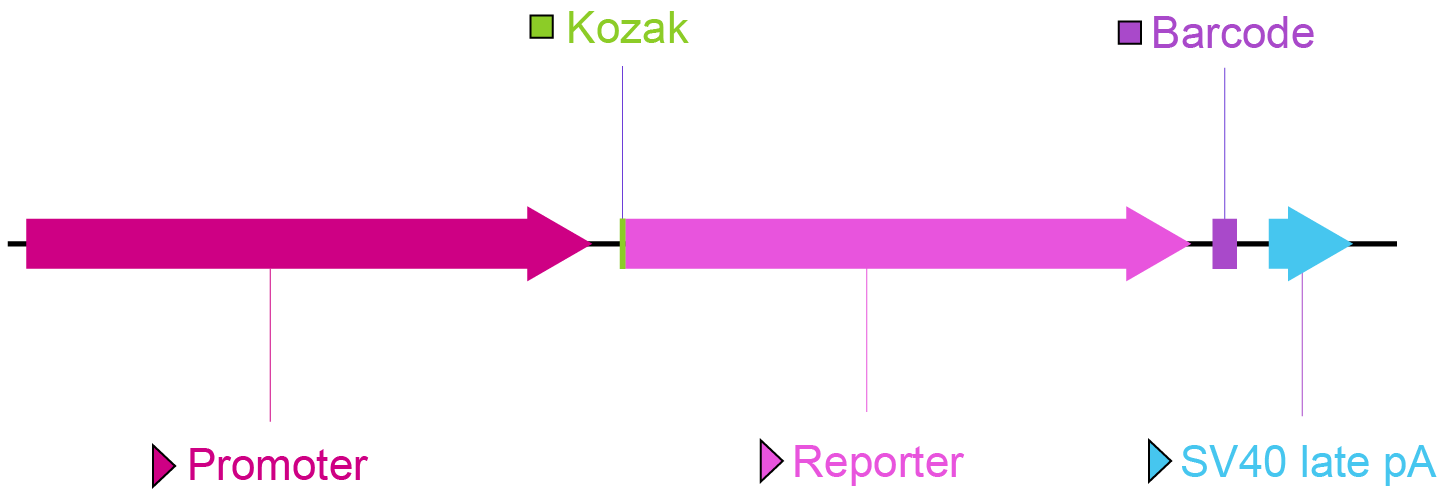
Figure 2. Key vector components for a promoter library.
Promoter: The promoter sequence to be screened can be inserted here to drive the transcription of the reporter.
Kozak: Kozak consensus sequence. It is placed in front of the start codon of the ORF of interest because it is believed to facilitate translation initiation in eukaryotes.
Reporter: A visually detectable bright fluorescent protein gene (such as GFP) or a chemiluminescent protein gene (such as luciferase). This allows highly sensitive detection of promoter activity.
Barcode: Promoter library barcodes are unique, short sequences within individual plasmids. After screening, the read count of each barcode is determined through NGS analysis. By correlating the unique barcode sequences with the specific promoter variants, the transcriptional activity of particular promoters can be identified.
SV40 late pA: Simian virus 40 late polyadenylation signal. It facilitates transcriptional termination of the upstream ORF.
An optimal library design is key to successful screening. Here are some important considerations for designing the enhancer/promoter library:
- Delivery system
While non-viral plasmids, lentivirus, and transposons are widely used for in vitro screening, AAV is often used for in vivo screening. It is crucial that the viral titer and serotype align with the administration route and tissue tropism. Additionally, when the library is high in complexity with a relatively low percentage of functional variants, non-viral plasmid transfection is typically favored over virus or transposon systems, as it results in a higher copy number of plasmids in cells, facilitating effective signal readout. For more information regarding the comparison of different vector systems, please visit our FAQ.
- Barcode
The incorporation of unique barcodes primarily depends on the type of library. When the enhancer/promoter is placed upstream of the reporter gene (Figures 1B and 2), a barcode should be positioned downstream of the reporter gene, before the 3' UTR. After screening, mRNA will be extracted, and the barcode—acting as a proxy for enhancer/promoter activity will be read using NGS, enabling the assessment of the transcriptional activity of the corresponding enhancer/promoter. Conversely, when the enhancer is positioned downstream of the reporter gene (Figure 1A), its activity can be measured directly through NGS. However, if the enhancer is lengthy, incorporating a barcode may still be necessary to ensure accurate readout. For further details on barcodes, please visit our Barcode Library Construction page.
- Scalability
Conducting cell sorting on target cells often enhances the effectiveness of detecting functional enhancers/promoters, thereby improving the scalability and efficiency of the screening process. Cell sorting can be achieved using known cell markers or by detecting the expression of the reporter gene from the library construct.
Library construction
The enhancer/promoter fragments can usually be obtained using common approaches mentioned below:
- Oligo pool synthesis
Chip-based oligo synthesis is ideal for generating pre-designed, short enhancer/promoter variants. This method can synthesize oligo pools up to 300 nucleotides long with a low error rate.
- Target enrichment from genomic DNA
Fragments from target genomic regions can be captured from size-selected, sheared genomic DNA or bacterial artificial chromosome (BAC) through probe hybridization. Alternatively, an unbiased shotgun approach can be utilized to include the entire genomic or BAC DNA.
- Directed evolution
Novel sequence variants can be developed from known enhancers/promoters using various mutagenesis strategies, including error-prone PCR, degenerate codon incorporation, or DNA shuffling.
In vitro/in vivo screening
The screening of enhancer/promoter variants can be conducted both in vitro and in vivo. In vitro selection is typically rapid and technically straightforward but fails to replicate the physiological conditions found in vivo. In vivo animal models provide a reliable platform for screening enhancers and promoters with improved transcriptional activity. Although both mice and non-human primates (NHPs) are utilized for in vivo selection of enhancer/promoter libraries, NHP models provide a more clinically relevant platform for identifying regulatory elements with desired properties due to their high degree of similarity to humans, a crucial factor in gene therapy applications.
For in vivo screens, multiple administration routes are available, including tail vein injection, facial vein injection (for neonatal mice and rats), intracerebroventricular injection, intrathecal injection, subretinal injection, intravitreal injection, intratympanic injection, intramuscular injection, and more.
After delivering the enhancer/promoter library to target cells or animals, the activity of candidate regulatory element variants can be assessed by measuring the expression of the reporter gene or barcode. To identify the regulatory elements with desirable properties, variants with enhanced activity can further undergo multiple rounds of screening. Figure 3 demonstrates the major steps for constructing a cone-specific promoter AAV library and in vivo screening.
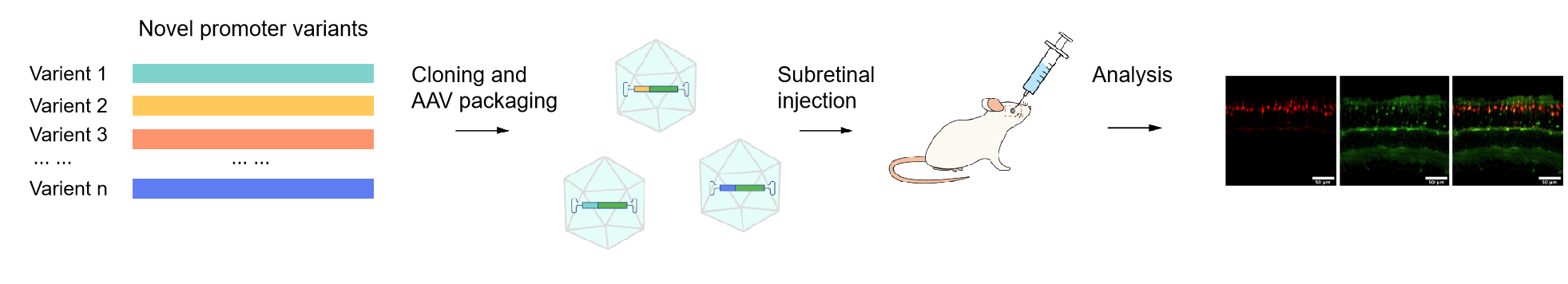
Figure 3. A cone-specific promoter AAV library construction and screening conducted by VectorBuilder. Novel promoter variants were generated, cloned into the AAV backbone, and used to drive reporter gene expression. The AAV promoter library was then injected into the subretinal region of mice, followed by comprehensive screening and analysis.
Hit validation
The positive hits identified from enhancer/promoter screens require further characterization for their desired activity, whether in vitro or in vivo. It is recommended to use the most functionally relevant system for characterization. For instance, in the case of the cone-specific promoter screens described in Figure 3, the identified novel promoter variants were characterized to determine their specificity and strength in mice and NHPs. Figure 4 below illustrates the functional validation of a novel VectorBuilder (VB) photoreceptor cone cell-specific promoter.
Figure 4. Functional characterization of a novel VB promoter identified from screening. (A) Cone-specific expression of nucleus-localized mCherry driven by the VB promoter in mice. AAV8 carrying the VB promoter driving mCherry was administrated by subretinal injection. Cone cells were labeled with the cone-cell marker Arrestin-C (green). (B) Cone-specific expression of mCherry driven by the VB promoter in crab-eating monkey. AAV8 carrying the VB promoter driving mCherry was administrated by subretinal injection.
How to Order
Customer-supplied library plasmid pool
If the customer-supplied premade plasmid pools are used, please send us the materials following the Materials Submission Guidelines. Please strictly follow our guidelines to set up shipment to avoid any delay or damage of materials. All customer-supplied materials undergo mandatory QC by VectorBuilder which may incur a $100 surcharge for each item. Please note that production may not be initiated until customer-supplied materials pass QC. For customer-supplied premade plasmid pools, we cannot provide any guarantees regarding the complexity or uniformity of the library.



