Related Services
Order a vector with your sequence in just a few clicks.Get plasmid preps of your sequence at a variety of scales.
Order high-titer packaging for ready-to-use viruses.

{[ account.info.display ]}
My Account
Codon optimization is a useful tool when expressing genes heterologously (in a different host organism), if problems occur when cloning a gene, or when optimizing gene expression level. VectorBuilder’s Codon Optimization tool is in-built into the Design Studio and can be used independently here.
Most amino acids can be translated from multiple codons, but codon bias reflects the preference for one codon over another and varies between species. This can lead to decreased translation when a gene is placed into a different host species. This tool can optimize the Codon Adaptation Index (CAI), taking advantage of the host organism’s codon bias to produce the same amino acid sequence at a higher efficiency.
VectorBuilder’s codon optimization tool is designed to help you achieve the optimal Codon Adaptation Index (CAI) for your GOI in any organism of your choice. It includes a comprehensive list of species and is seamlessly incorporated into our online vector design platform enabling you to optimize your GOIs while designing vectors. Additionally, it allows you to avoid cleavage sites of selected restriction enzymes while codon optimizing your target sequence. Our tool can be used for optimizing sequences with extreme GC content and simple repeats for highly efficient gene synthesis and DNA cloning applications.
Codon optimization can additionally be used to enhance cloning efficiency of a gene of interest. This tool can aid in optimizing GC content and repetitive sequences, improving mRNA stability, and avoiding restriction enzyme recognition sites, thus improving transcription or translation efficiency. At any time you can design a vector containing your sequence of interest and order downstream services.
Below are some examples illustrating various functionalities of our codon optimization tool:
Figure 1 below illustrates that when the sequence of native piggyBac transposase from Trichoplusia ni was optimized for expression in human using our tool, it resulted in an optimized sequence with a CAI of 0.93. The CAI of the piggyBac transposase gene prior to optimization was found to be 0.69. The CAI for a species is a quantification of the frequency of usage of favored codons in highly expressed genes. CAI values range between 0 and 1. The higher the CAI value of a gene for a specific target species, the greater the chances are for it to be expressed optimally in that species.
Figure 1. Optimizing a sequence for codon usage in a target species using VectorBuilder’s codon optimization tool.
Figure 2 illustrates that when the mouse Hoxa4 gene with an overall GC content of 69.3% was optimized using our tool, it resulted in the GC content to drop to 59.5%. For genes requiring synthesis during the cloning process, an optimal GC content of approximately 60% is recommended to increase the chances for the gene synthesis to succeed.
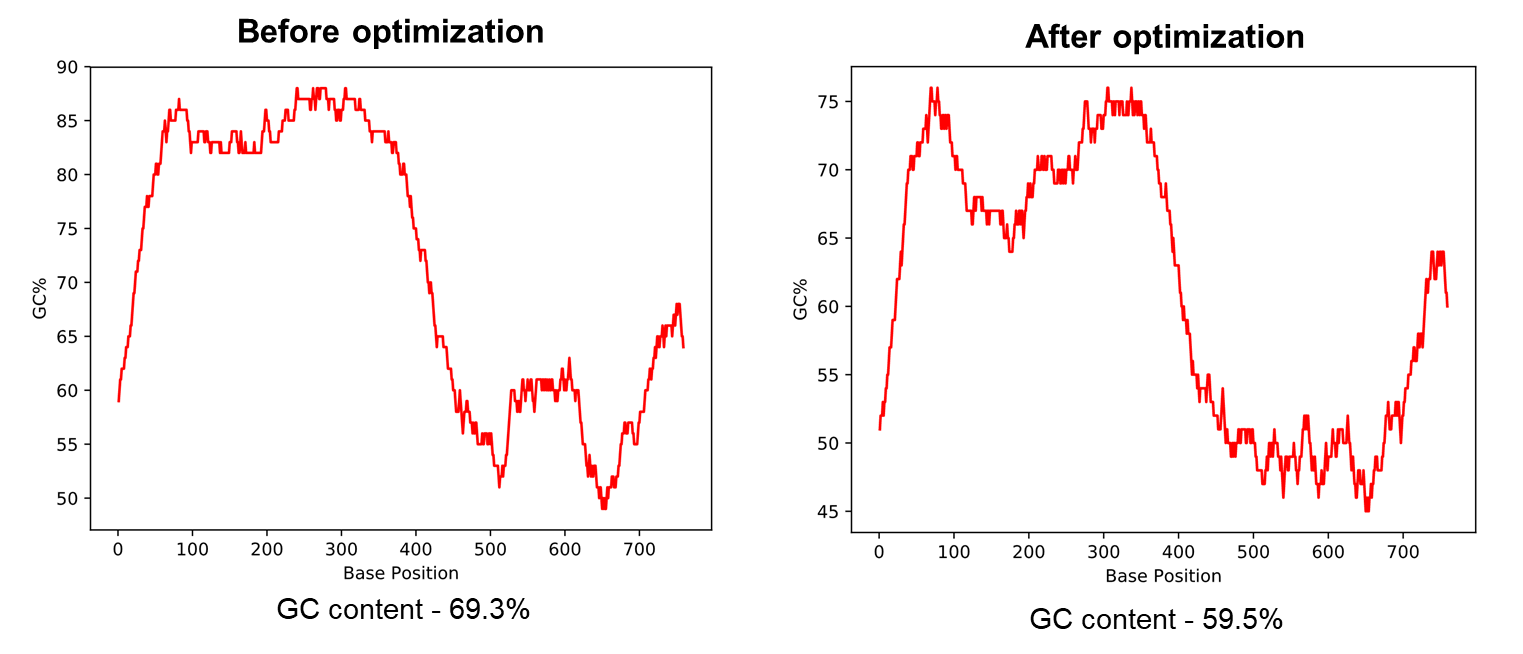
Figure 2. Optimizing a sequence with high GC content using VectorBuilder’s codon optimization tool.
Figure 3 below illustrates dot plots comparing the human immunoglobulin heavy chain sequence against itself, before and after codon optimization with our tool. While the before optimization dot plot shows the presence of highly repetitive regions within the sequence indicated by the multiple diagonal lines, optimization of the sequence resulted in a significant decrease in the repeats, as shown by the after optimization dot plot.
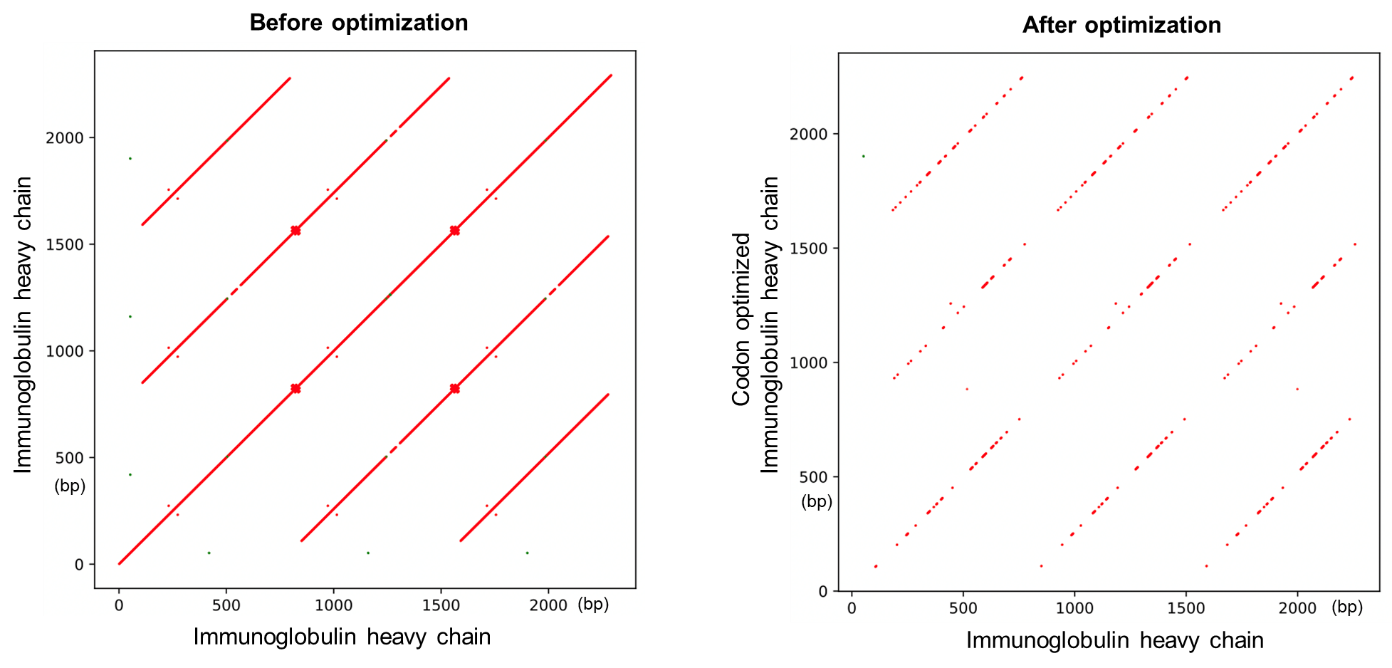
Figure 3. Optimizing a sequence for reducing repetitive regions using VectorBuilder' s codon optimization tool.
In order to produce proteins, a cell must first translate the relevant mRNA strand. Following transcription, the mRNA exits the nucleus where each group of three nucleotides is matched to a tRNA molecule carrying an amino acid (Figure 1A). These groups of 3 nucleotides are codons, and each corresponds to an amino acid. Because there are only 20 amino acids and many more possible combinations of nucleotides, there is redundancy in this code (Figure 1B).


Figure 1. Formation of a protein through transcription and translation (A) of codons. Each codon corresponds to an amino acid or direction (start/stop).
Although there are multiple options for making each amino acid, their usage is not based on chance. This is because each species exhibits codon bias, the preference for making an amino acid with a certain codon. For instance, alanine (Ala) is coded by GCU, GCC, GCA, and GCG (Figure 1B), but in humans, GCC is used about 40% of the time. Different organisms have different codon preferences, which influences RNA processing and therefore protein folding and function. This creates complications when expressing one gene in another organism, i.e. heterologous gene expression.
The Codon Adaptation Index (CAI) is a measure of how well given codons match with the biases of an organism, ranging from 0 to 1. A CAI of 1 reflects a coding sequence where all amino acids reflect the most frequently used codons in that organism. Our Codon Optimization tool presents a sequence that balances an optimal CAI with other factors that can influence molecular experiments.
Codon optimization can also aid in increasing cloning efficiency based on the distribution of nucleotides across the sequence. GC content is an important variable to consider when designing and troubleshooting experiments. If GC content is too high or too low, stability of the query sequence is negatively affected. Our GC Content Calculator tool allows for independent GC analysis over an entire sequence and within segments of a sequence. However, our Codon Optimization tool incorporates this analysis to optimize this variable by finding synonymous codons that increase or decrease GC content as needed.
Additionally, sequences that have a high frequency of repeats can present complications in cloning efforts due to the lack of unique primer binding sites, and sequences with recognition sites for restriction enzymes can present challenges in experimental design. Using our Codon Optimization tool allows for all of these factors to be optimized in unison with codon bias to provide a sequence that is most likely to efficiently produce your protein in your system.
