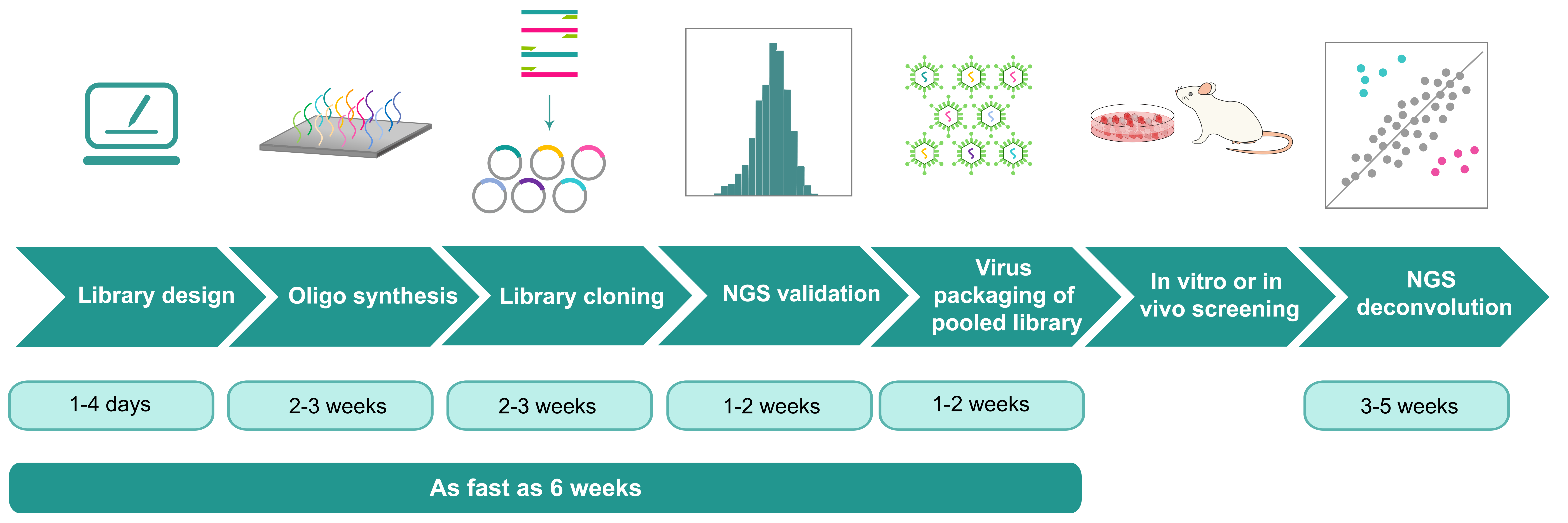
shRNA Library Screening
VectorBuilder excels in delivering high-quality, custom pooled RNAi knockdown libraries, offering a potent and cost-effective solution for large-scale loss-of-function screens. Our shRNA libraries can be delivered in various formats, including E. coli stock, plasmid DNA, or recombinant virus, and each library undergoes thorough validation through next-generation sequencing (NGS). We further offer both in vitro and in vivo pooled screening of shRNA libraries to accommodate a wide range of research demands.
Highlights
- One-stop library shop: From design to construction and screening to deconvolution, VectorBuilder provides any and all library services.
- Wide range of complexity: We can generate shRNA libraries with complexity ranging from a few hundred to up to 106.
- Unlimited customization of vector design: We can fully customize the vector design based on your requirements on delivery system, promoter, marker, barcode, and more.
- High-quality virus packaging: We can do packaging for various viral systems with fast turnaround and affordable prices.
- Great coverage and high uniformity: Our libraries typically achieve >98% coverage of designed variants.
Service Details
Price and turnaround Price Match
Table 1. Price and turnaround by service module
| Service Module | Brief Description | Price (USD) | Turnaround |
|---|---|---|---|
| Library design | Vector backbone design for shRNA knockdown screening, and shRNA sequence design for your target genes. | Free | 1-4 days |
| Pooled library cloning (including oligo synthesis and NGS validation) | Includes shRNA synthesis, massive parallel cloning of shRNA into desired vector backbone, preliminary validation by Sanger sequencing, and full validation by NGS. Deliverable includes library in E. coli glycerol stock and NGS report. See Table 2 below for details. | ||
| Virus packaging of pooled library | Please click here to view detailed information of virus packaging services. The price for packaging library plasmid is 1.5-fold of the price for packaging single vector plasmid. | ||
| Pooled screening of shRNA library | In vitro or in vivo screening of your libraries. Includes cell viability, drug resistance, pathway, reporter, phenotypic, target-based, and toxicity screening, as well as other custom screenings | Please inquire |
Project specific |
| NGS deconvolution of post-screening sample | Includes NGS library preparation from genomic DNA of screened cells, Illumina sequencing (>500x coverage), and data analysis. | From $320 per sample | 3-5 weeks |
Table 2. Library cloning price by library complexity
| Library Complexity | Price (USD) | Turnaround |
|---|---|---|
| <5x103 | From $2,800 | 5-8 weeks |
| 5x103 ~ 1x104 | From $5,000 | |
| 1x104 ~ 5x104 | From $6,000 | 6-10 weeks |
| 5x104 ~ 1x105 | From $8,000 | |
| >105 | Please inquire | |
Technical Information
Workflow of pooled shRNA library-mediated genetic screens
A conventional workflow of genetic screens mediated by pooled shRNA lentivirus libraries is shown in Figure 1 below. First, cells of interest are transduced with the shRNA lentivirus library, and positively transduced cells are selected by the marker gene(s) carried on the lentiviral genome (e.g. drug-selection or fluorescence marker). Next, positively transduced cells are split into reference population and experimental population. Then, the experimental population is subjected to particular selective pressure (e.g. drug treatment or repeated passaging) to identify cells with the phenotype(s) of interest. There are three major types of screening strategies: 1) viability screens that search for shRNAs enriched or depleted in surviving cells when exposed to selective pressure; 2) reporter screens that look for shRNAs enriched in cells associated with either high or low reporter expression (e.g. shRNAs targeting transcription factors that modulate reporter gene expression); 3) behavior screens that usually identify shRNAs affecting genes associated with cell invasion, migration, etc. After screening, both experimental and reference cells are harvested, and enriched or depleted shRNAs in the experimental group compared to the reference group are identified using Sanger sequencing or next-generation sequencing. Candidate genes that are potentially targeted by the enriched or depleted shRNAs can be further investigated by downstream functional studies.
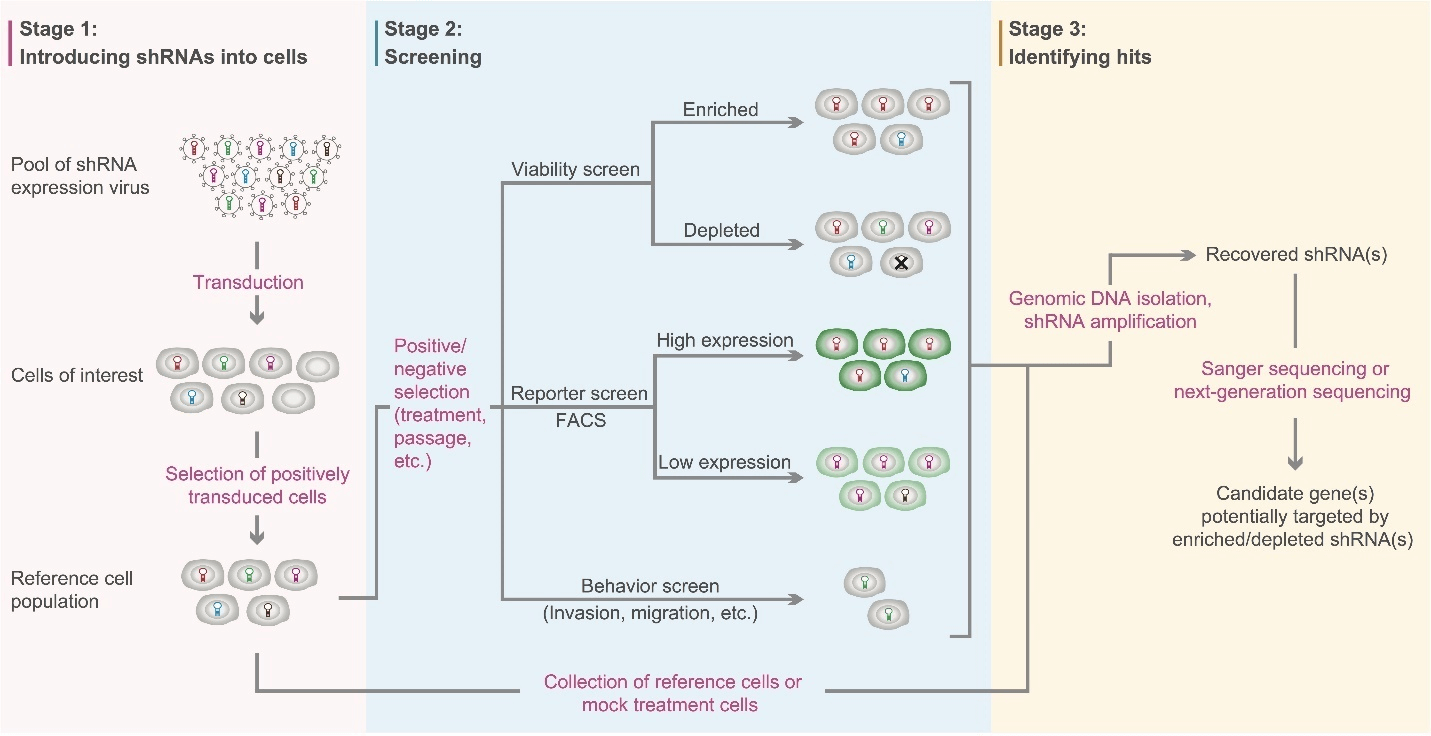
Figure 1. Workflow of loss-of-function screens mediated by pooled shRNA lentivirus libraries. Adapted from Acta Biochim Biophys Sin 44:103-112 (2012).
Experimental data
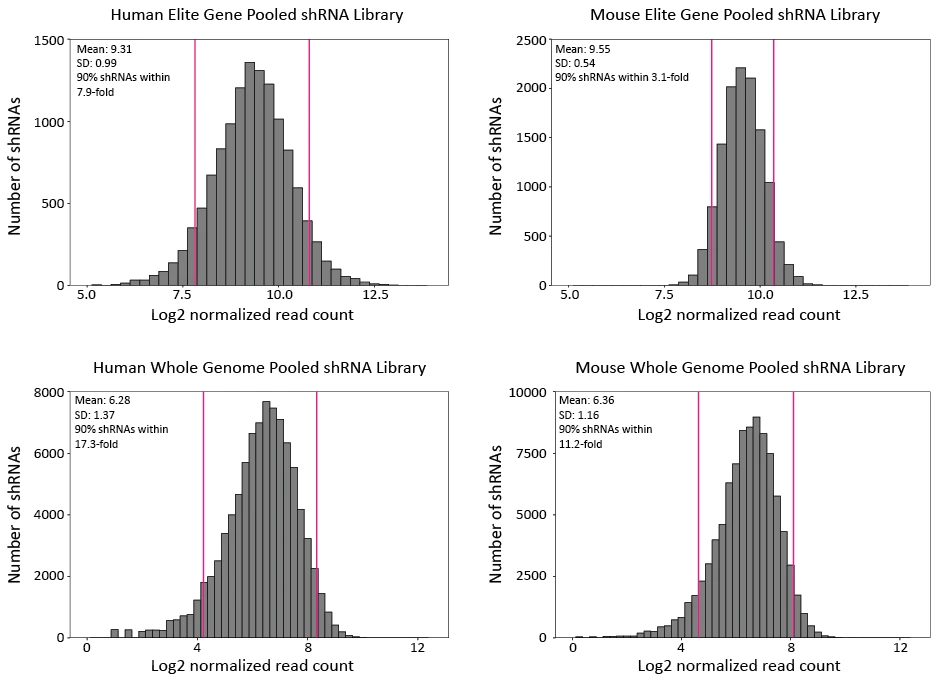
Figure 2. Distribution of shRNA representation in human and mouse elite gene (~2,000 most frequently cited genes on PubMed Central) and whole genome pooled libraries. shRNA read counts were normalized by NGS library size (10 million reads) and plotted in log2 scale.
How to Order
Customer-supplied library plasmid pool
If the customer-supplied premade plasmid pools are used, please send us the materials following the Materials Submission Guidelines. Please strictly follow our guidelines to set up shipment to avoid any delay or damage of materials. All customer-supplied materials undergo mandatory QC by VectorBuilder which may incur a $100 surcharge for each item. Please note that production may not be initiated until customer-supplied materials pass QC. For customer-supplied premade plasmid pools, we cannot provide any guarantees regarding the complexity or uniformity of the library.
Resources
FAQ
| Array-based Screen | Pooled RNAi Screen | |
|---|---|---|
| How are shRNAs introduced into cells of interest? | Individual shRNAs are applied to cells grown in different wells across multi-well plates (e.g. 96-well or 384-well). | Hundreds and thousands of different shRNAs are applied to the cell population simultaneously. |
| How to identify shRNAs associated with phenotypes of interest? | Wells showing phenotypes of interest are selected by examining phenotypes well-by-well. The identity of the shRNAs applied to individual wells are already known. | Cells with phenotypes of interest are selected from the population. shRNAs enriched/depleted in cells of interest are identified by sequencing. |
| Can genetic interactions be detected? | No (if a single shRNA is added to each well) or limited (if more than one shRNAs are added to each well). | Yes (cells may carry multiple randomly combined shRNAs). |
| Technical variation | High | Low |
| Cost of labor and reagents | High | Low |
| Requirement of special equipment | High (e.g. liquid handler, high-throughput imaging, etc.) | Low (conventional benchtop equipment) |
VectorBuilder applies rules similar to that used by the RNAi consortium (TRC) to design and score shRNAs. For each given RefSeq transcript, we search for all possible 21-mers that are considered as candidate target sites. Candidates are excluded if they contain features thought to reduce knockdown efficiency/specificity or clonability, including a run of ≥4 of the same base, a run of ≥7 G or C, GC content <25% or >60%, and AA at the 5’ end. Knockdown scores are penalized for candidates that contain internal stem-loop, high GC content toward the 3’ end, known miRNA seed sequences, or off-target matches to other genes. For genes with alternative transcripts, target sites that exist in all transcripts are given higher scores.
All scores are ≥0, with mean at ~5, standard deviation at ~5, and 95% of scores ≤15. A shRNA with a knockdown score of about 15 is considered to have the best knockdown performance and clonability, while an shRNA with a knockdown score of 0 has the worst knockdown performance or is hard to be cloned.
Please note that knockdown scores are only a rough guide. Actual knockdown efficiency could depart significantly from what the scores predict. Target sites with low scores may still work well. Also, please note that targeting 3’ UTR can be as effective as targeting coding region.