AAV Capsid Evolution
Recombinant adeno-associated virus (AAV) is a highly popular gene delivery vector for a wide range of gene therapy and vaccine applications, thanks to its broad tropism, prolonged transgene expression, non-pathogenicity, and low immunogenicity.
However, several problems with existing AAV serotypes limit their therapeutic potential. First, while available serotypes offer a variety of tropisms to choose from, many clinical applications require tissue-specificities that fall outside of their coverage. Second, even if a tissue of therapeutic interest is covered by the tropism of one or more serotypes, the efficiency of gene delivery may be too low and there may also be undesirable tropism for off-target tissues. Third, pre-existing neutralizing antibodies against many AAV serotypes can block their efficient delivery initially or with repeated drug administration. Lastly, some serotypes are inherently difficult to manufacture at high titer, purity and stability. To overcome these limitations, AAV capsid engineering has been a critical area of research that has significantly accelerated the development of novel AAV variants with improved features.
Directed evolution is a widely used high-throughput approach to engineer enhanced biomolecules. It mimics the process of natural selection through repeated genetic diversification and selection. Directed evolution of AAV capsid is performed by mutating the wildtype AAV capsid gene to generate highly diverse AAV capsid libraries, which are then screened to identify novel capsid variants with improved properties. Since directed evolution does not require prior knowledge of the structure-function relationship of proteins, it is often preferred over rational design for AAV capsid engineering.
Service Highlights
- Full-service platform: VectorBuilder is the world’s only company capable of providing one-stop solution for all your AAV preclinical and clinical CRO and CDMO needs. Our AAV services encompass vector design and optimization, vector cloning, library construction, virus packaging, capsid evolution and targeted engineering, AAV biodistribution profiling, and GMP manufacturing.
- High complexity capsid library construction via versatile approaches: With our extensive expertise in constructing pooled libraries, we can help you design and custom build highly diverse capsid libraries via any mutagenesis approach or combinatorial approach.
- High titer capsid library virus packaging: We can do either one-step or two-step virus packaging of the capsid library to ensure each viral particle to contain corresponding capsid variant in its genome and to achieve high virus titer.
- In vivo screening in multiple species including nonhuman primate (NHP): Besides in vitro screening capabilities, we offer a wide range of species on our in vivo screening platform, such as mouse, rat, and importantly, two nonhuman primate species: crab-eating macaque (Macaca fascicularis; a.k.a. cynomolgus monkey) and rhesus macaque (Macaca Mulatta). In vivo screenings are carried out in AAALAC-accredited facilities by highly trained professionals.
- Full technical support: Our highly experienced scientists can provide comprehensive technical support covering every aspect of your AAV capsid project, ranging from library construction to in vivo screening, from library design to NGS analysis.
Workflow of Capsid Evolution and Screening
The first and the most critical step in the entire workflow of AAV capsid evolution is the generation of a highly diverse AAV capsid library in which each plasmid carries a chimeric AAV genome consisting of a rep gene and a capsid gene variant. The capsid gene variants can be efficiently generated using various approaches, such as error-prone PCR, random peptide display, DNA family shuffling, or in silico design. The capsid library is then packaged into viral particles, and each viral particle harbors corresponding capsid variant in its genome. The viral library is subsequently subjected to a screening process to test the ability of chimeric AAVs: 1) to efficiently transduce target cells within specific tissues or organs, or 2) to bind to cell type-specific receptors at high affinity, or 3) to evade neutralizing antibodies. Viral genomes passed screening are recovered from target cells and made into a smaller library for a second round of screening. Several rounds of screening are usually performed to enrich high confidence hits. The resulting hits are then validated and characterized to identify novel AAV capsid variants with enhanced properties (Figure 1).
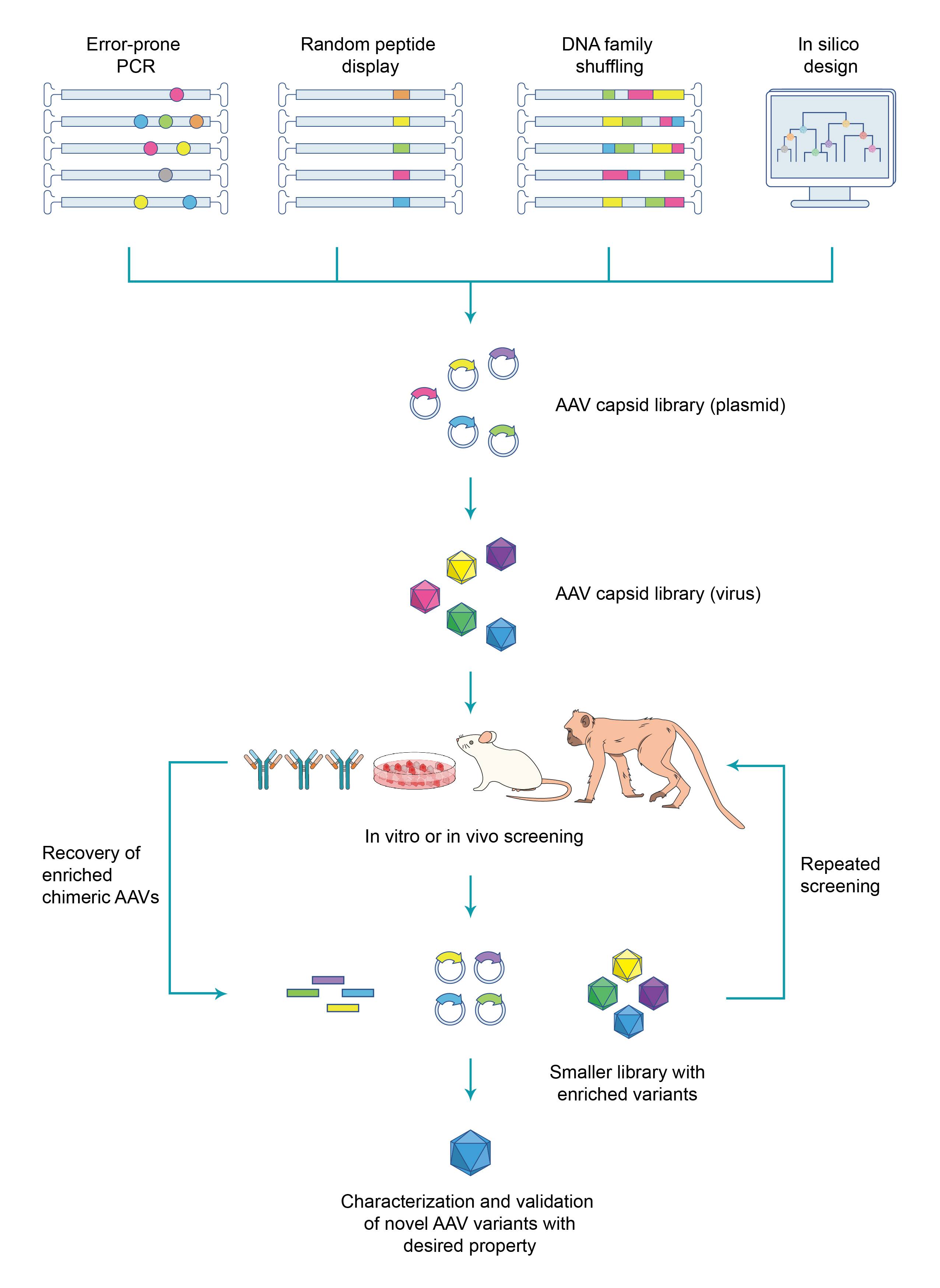
Figure 1. Typical workflow for screening novel AAV capsids by directed evolution.
Capsid library construction
The strategies described below are commonly used for creating diverse AAV capsid variants. Then, capsid library for screening is constructed by massive parallel cloning of the capsid variants into AAV vector to form chimeric AAV genomes (each consisting of a rep gene and a capsid gene variant).
Error-prone PCR
Error-prone PCR is the most straightforward approach for developing highly variable AAV capsid libraries which involves the modification of standard PCR methods to mutagenize the AAV capsid gene. More specifically, error-prone PCR employs a combination of various sub-optimal PCR conditions including low fidelity polymerases, longer extension times, higher Mg2+ concentrations, addition of Mn2+, and varying dNTP concentrations for introducing random point mutations into the AAV capsid gene.
Random peptide display
Random peptide display involves the insertion of random peptide sequences of usually 7 to 9 amino acids into specific sites of the AAV capsid with the aim of altering the natural cellular interactions of the virus and retargeting it to specific cell receptors. The peptides are typically inserted into locations of the AAV capsid that facilitate surface exposure of the peptide and are also critical for virus-host interactions. For example, between position 587 and 588 (within the variable region VRIII) of the AAV2 capsid is a preferred insertion site for most AAV2-based peptide display libraries since insertion of peptides in that region abolishes the heparan sulfate proteoglycan (HSPG, the primary AAV2 receptor) motif of AAV2 and enables the displayed peptides to interact efficiently with cell surface molecules.
DNA family shuffling
DNA family shuffling is a highly efficient approach for generating chimeric AAV capsids by molecular interbreeding of parental capsid genes derived from different AAV serotypes. To accomplish this, the parental capsid genes of various AAV serotypes are fragmented, followed by their reassembly into novel full-length capsid variants by primer-less PCR which recombines them based on partial sequence homology. As an alternative strategy, high complexity libraries can also be created by synthetic shuffling which combines rational design (modifying the capsid based on prior knowledge of AAV biology) with directed evolution. In this approach, capsid locations suitable for mutagenesis are first identified and evaluated based on a detailed structural and sequence analysis of naturally occurring AAV serotypes. Fragments containing mutations are synthesized and assembled to form full-length novel capsid variants.
In silico design
In silico design of AAV capsid libraries utilizes a variety of approaches for computational prediction of capsid variant sequences with the potential to contribute to enhanced AAV performance. One commonly used approach is ancestral reconstruction which involves in silico designing of a putative ancestral AAV library followed by its experimental validation for identifying highly potent ancestral capsid sequences with improved tropism. The rationale behind this approach is that evolutionary AAV intermediates that emerged by surviving the process of natural selection are highly likely to possess unique properties while maintaining virus structure and function. Machine learning is another commonly used in silico design approach that applies computational algorithms to predict the chances of viable virus production from hypothetical capsid variants. Machine learning algorithms heavily rely on available input data to learn structure-function relationships of proteins and apply that to predict the outcome of complex physiological processes such as viral capsid assembly.
Virus packaging of capsid library
Virus packaging of capsid library can be accomplished by either one- or two-step process as described below:
One-step packaging of capsid library
The conventional approach for packaging AAV capsid libraries utilizes a one-step process in which packaging cells are co-transfected with capsid library and an adenoviral helper plasmid. While this approach is widely used, it does come with the disadvantages of cross-packaging (generation of AAV particles with mismatched capsid variant genome and capsid) and capsid mosaicism (generation of AAV particles with mosaic capsids arising from capsid proteins derived from different genomes). To overcome these challenges, it is recommended that the packaging cells are transfected at a very low plasmid library to cell ratio to ensure the uptake of at most one single library plasmid per cell.
Two-step packaging of capsid library
In a two-step packaging approach, the capsid library is first co-transfected into packaging cells along with a helper plasmid encoding the WT capsid gene but lacking the viral ITRs. This results in the production of AAV particles with mosaic capsids that are partially made up of the WT capsid and are referred to as AAV transfer shuttles. The AAV transfer shuttles are then introduced into packaging cells at a low MOI for achieving at most one viral particle per cell, followed by superinfection of the packaging cells with adenovirus which ultimately results in the generation of high-titer viral capsid library.
Capsid screening in vitro and in vivo
The viral capsid library is usually subjected to several rounds of screening in vitro or in vivo, based on screening purpose, to select for chimeric AAVs with desirable properties:
In vitro selection
Utilizing established cells lines for AAV capsid library selection is a widely used approach, particularly for identifying AAV variants with altered receptor targeting abilities. Although in vitro selection of AAV libraries is fast and technically simple, it does present some challenges. Firstly, vectors optimized for high transduction efficiency in vitro may not be able recapitulate the same efficiency when used in vivo. Secondly, AAV vectors demonstrating high degree of target cell specificity in vitro might transduce non-target tissues when translated in vivo. Another strategy for in vitro selection of AAV libraries involves subjecting the library to potent serum from immunized animals prior to adding it to target cells, specifically for identifying variants with immune evasive properties. However, immune response of AAV variants may vary when translated in vivo due to various factors (e.g. immune recognition of the same AAV vector may vary when delivered via different routes).
In vivo selection
In vivo animal models offer a more reliable platform for screening AAV libraries, particularly for identifying AAV variants capable of transducing delicate cell types that cannot be grown in culture or AAV variants capable of transducing a specific cell type with a complex tissue structure. In vivo selection also helps to identify any potential off-target effects associated with an AAV variant. While both mouse and NHPs are widely used for in vivo selection of AAV libraries, NHP models represent the most clinically relevant platform for screening improved AAV vectors due to their high degree of similarity to humans.




