Library Screening
VectorBuilder is your full-service platform for library construction and screening services, streamlining your research from start to finish. Our team of experts is dedicated to assisting you in experimental design, creating high-quality libraries, and performing library screens that are tailored to your specific research goals. We ensure the highest quality throughout the experimental process, from construction of NGS-validated pooled libraries to efficient deconvolution.
Highlights

One-stop shop
Optimized workflow from experimental design through library construction to screening and analysis, guided by our scientific team's unparalleled expertise in cloning and library preparation.
Customization
Tailor-made services designed to meet unique research needs, speed up timelines and improve results.

Extensive experience
Specializing in the creation of various types of high quality, high-complexity libraries with extensive coverage across a variety of genomes.

Comprehensive screening solutions
Offering both in vitro and in vivo screening to meet the broadest range of research demands.
Custom Libraries
Premade Pooled Libraries
VectorBuilder can provide library services from start to finish or standalone offerings, providing you with exactly the service and library screen you need.
Order Now!Workflow for Library Construction

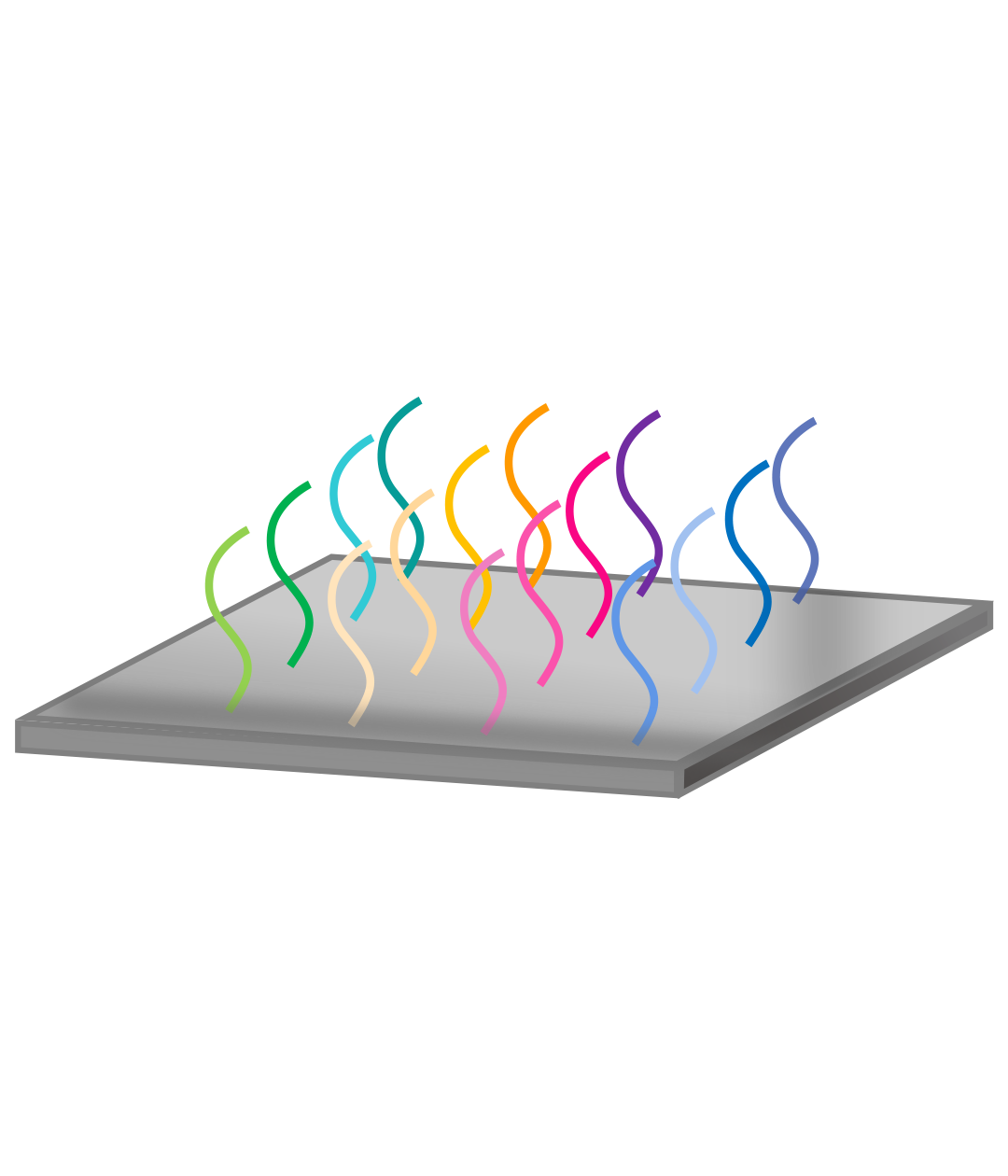
Library design
Oligo synthesis
Library cloning
NGS validation
Virus
packaging of
pooled library
packaging of
pooled library
In vitro or in
vivo screening
vivo screening
NGS
deconvolution
deconvolution

Library design
Designing an optimized library is critical to a successful screening experiment. Our technical team can help you design a library vector that is compatible with an efficient and error-less cloning process, contains proper marker or barcode sequence for versatile downstream screening experiments, and has high signal-to-noise ratio for robust results. Depending on your research needs, you can choose from our various validated delivery systems, such as lentivirus, AAV, piggyBac, non-viral regular plasmid, and adenovirus.
For CRISPR or RNAi screen, if you need assistance in designing gRNA or shRNA sequences for your list of target genes or genomic regions, VectorBuilder can help you design them using our established algorithm and computational pipeline. For 2-vector CRISPR libraries, we can also construct and package Cas9 expression vectors that are suitable for your screening strategy.
Pooled library cloning
To obtain DNA variants to be inserted in the vector backbone, we can use de novo oligo synthesis, directed evolution, or combinatorial approach. The variants are PCR amplified for a minimal number of cycles and in-parallel cloned into the desired vector backbone with high coverage. E. coli cells harboring library plasmids are propagated and harvested in exponential growth phase to make glycerol stocks. To assess the coverage and uniformity of the library, NGS analysis is performed on the plasmid pool before proceeding to virus packaging.
Virus packaging of pooled library
You may need to deliver the pooled library into cells via viral transduction, such as infecting with lentivirus or AAV. VectorBuilder has developed a range of proprietary technologies and reagents that have greatly improved virus packaging protocols for pooled libraries in terms of titer, purity, and uniformity. We can deliver the ready-to-use viral libraries for your screening experiments with a fast turnaround.
Click to view more information of virus packaging servicesIn vitro or in vivo screening
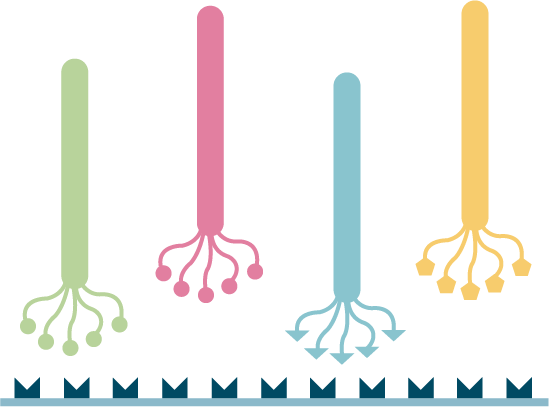
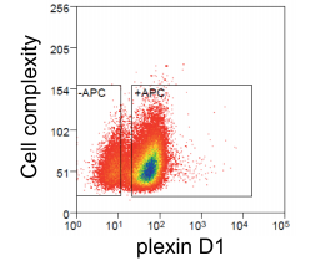
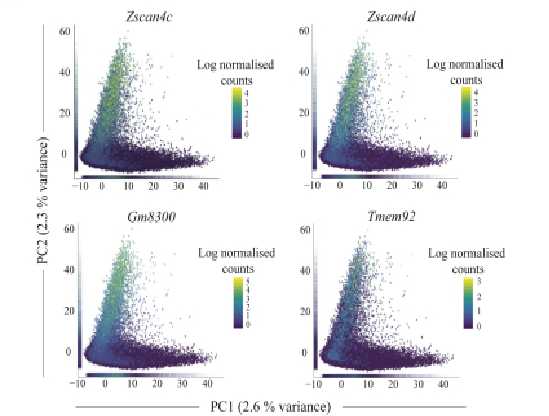
High-throughput library screening utilizes analytical methods to efficiently sift through vast libraries for a wide range of applications including gene function analysis, target identification for drug development, pathway analysis, and detection of genetic markers and phenotypes. Our technical team can help you with a diverse range of pooled screening options both in cells and in vivo tailored to meet your specific research needs, including cell viability/survival screening, drug resistance screening, pathway screening, target-based screening, toxicity screening, and custom screenings designed around your specific research objectives.
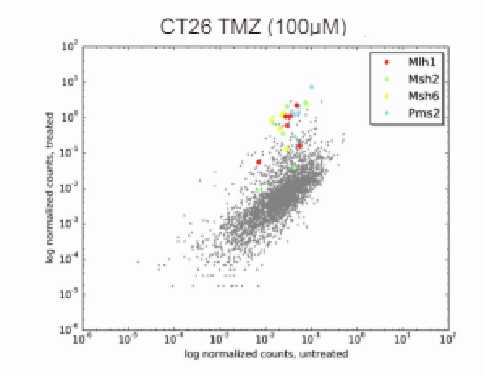
NGS deconvolution
VectorBuilder can help you identify hits by NGS for your pooled library screens. You can simply send the genomic DNA of your samples to VectorBuilder, and we will prepare the NGS libraries, perform high-throughput sequencing, analyze the data, and deliver the accurate read counts in each sample to you in just a few weeks.
Service Details
Price and turnaround Price Match
| Service Module | Brief Description | Price (USD) | Turnaround |
|---|---|---|---|
| Library design | Library vector backbone design for CRISPR, RNAi, barcode, enhancer/promoter, mutation, AAV capsid, and more pooled libraries, and gRNA or shRNA sequence design for your target genes/regions. | Free | 1-4 days |
| Pooled library cloning (including oligo synthesis and NGS validation) | Massive parallel cloning of DNA variants into desired vector backbone, followed by preliminary QC by Sanger sequencing and full QC by NGS. The default deliverable is E. coli glycerol stock. | From $2,300 | 5-8 weeks |
| Virus packaging of pooled library | Please click here to view detailed info of virus packaging services. The price for packaging library is 1.5-fold of the price for packaging single vector. | ||
| Pooled screening of library samples | In vitro or in vivo screening of your libraries. Includes cell viability screening, drug resistance screening, pathway screening, reporter screens, phenotypic screening, target-based screening, toxicity screening, and other custom screenings | Please inquire | Project specific |
| NGS deconvolution of post-screening sample | Includes NGS library preparation from genomic DNA of screened cells, Illumina sequencing (>500x coverage), and data analysis. | From $320 per sample | 3-5 weeks |
How to Order
Customer-supplied library plasmid pool
If the customer-supplied premade plasmid pools are used, please send us the materials following the Materials Submission Guidelines. Please strictly follow our guidelines to set up shipment to avoid any delay or damage of materials. All customer-supplied materials undergo mandatory QC by VectorBuilder which may incur a $100 surcharge for each item. Please note that production may not be initiated until customer-supplied materials pass QC. For customer-supplied premade plasmid pools, we cannot provide any guarantees regarding the complexity or uniformity of the library.
VectorBuilder can provide library construction or library screening alone, or we can provide a combination of library construction and screening services
Order Now!