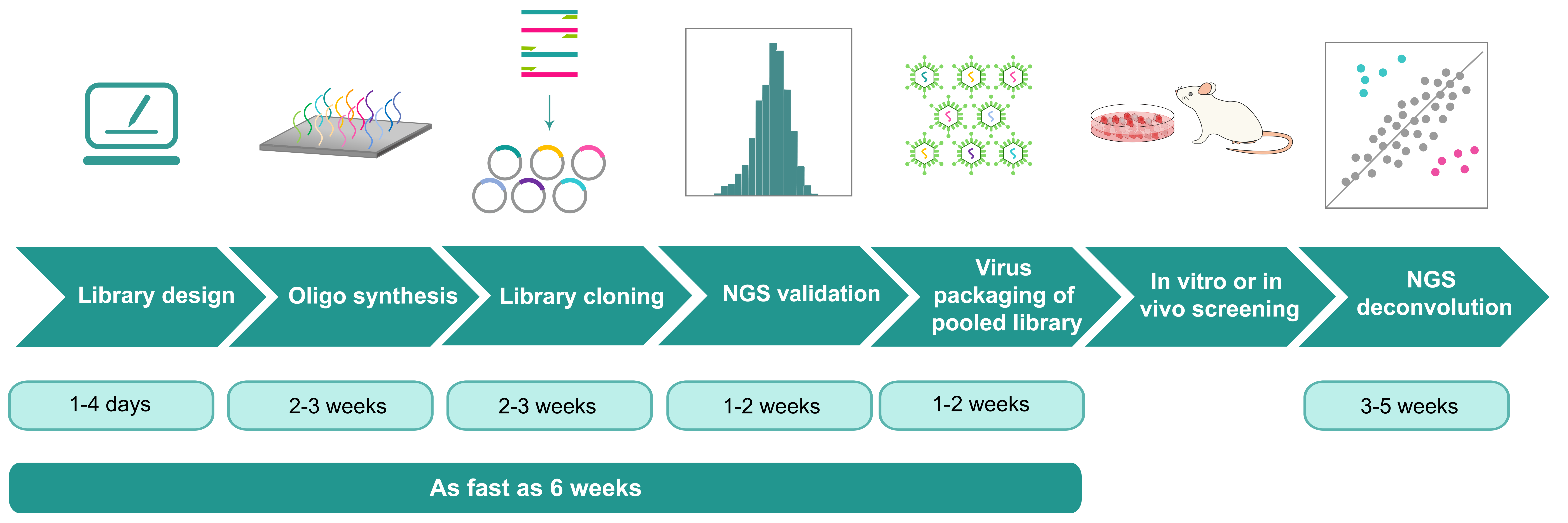
CRISPR Library Screening
VectorBuilder specializes in crafting high-quality CRISPR libraries and performing comprehensive large-scale functional screens tailored to meet diverse needs. Our CRISPR libraries can be produced in various formats, including E. coli stock, plasmid DNA, or recombinant virus, with thorough validation of each library through next-generation sequencing (NGS). We further offer both in vitro and in vivo pooled screening of CRISPR libraries to accommodate a wide range of research demands.
Highlights
- One-stop library shop: From design to construction and screening to deconvolution, VectorBuilder provides any and all library services.
- Wide range of complexity: We can generate CRISPR libraries with complexity ranging from a few hundred to up to 106.
- Experience in various CRISPR technologies: We have extensive experience in building libraries for all kinds of screening technologies, such as CRISPR knockout (single or dual gRNA), CRISPRa, CRISPRi, single-cell CRISPR technologies, CRISPR barcoding technologies, and more.
- Unlimited customization of vector design: We can fully customize the vector design based on your requirements on delivery system, promoter, marker, barcode, and more.
- High-quality virus packaging: We can do packaging for various viral systems with fast turnaround and affordable price.
- Great coverage and high uniformity: Our libraries typically achieve >98% coverage of designed gRNAs.
Service Details
Price and turnaround Price Match
Table 1. Price and turnaround by service module
| Service Module | Brief Description | Price (USD) | Turnaround |
|---|---|---|---|
| Library design | Vector backbone design for CRISPR screening, and gRNA sequence design for your target genes/regions. | Free | 1-4 days |
| Pooled library cloning (including oligo synthesis and NGS validation) | Includes guide RNA synthesis, massive parallel cloning of gRNA into desired vector backbone, preliminary validation by Sanger sequencing, and full validation by NGS. Deliverable includes library in E. coli glycerol stock and NGS report. See Table 2 below for details. | ||
| Virus packaging of pooled library | Please click here to view detailed info of virus packaging services. The price for packaging library plasmid is 1.5-fold of the price for packaging single vector plasmid. | ||
|
Pooled screening of CRISPR library |
In vitro or in vivo screening of your libraries. Includes cell viability, drug resistance, pathway, reporter, phenotypic, target-based, and toxicity screening, as well as other custom screenings | Please inquire | Project specific |
| NGS deconvolution of post-screening sample | Includes NGS library preparation from genomic DNA of screened cells, Illumina sequencing (>500x coverage), and data analysis. | From $320 per sample | 3-5 weeks |
Table 2. Library cloning price by library complexity
| Library Complexity | Price (USD) | Turnaround |
|---|---|---|
| Single-gRNA | ||
| <5x103 | From $2,300 | 5-8 weeks |
| 5x103 ~ 1x104 | From $4,500 | |
| 1x104 ~ 5x104 | From $5,500 | 6-10 weeks |
| 5x104 ~ 1x105 | From $7,500 | |
| >1x105 | Please inquire | |
| Dual-gRNA | ||
| <5x103 | From $3,300 | 9-13 weeks |
| 5x103 ~ 1x104 | From $6,500 | |
| 1x104 ~ 5x104 | From $8,000 | |
| 5x104 ~ 1x105 | From $10,000 | |
| >1x105 | Please inquire | |
Technical Information
CRISPR screening strategies
Pooled CRISPR libraries are powerful tools for performing large-scale or genome-wide functional screens of coding or noncoding regions involved in particular pathways, diseases, cell responses to drug treatment, developmental processes, gene regulation, etc. Table 3 below lists and compares commonly used CRISPR-based screens that VectorBuilder can perform:
Table 3. Comparison of CRISPR screening approaches
| Screening Strategy | Mechanism | Target Location | Application |
|---|---|---|---|
| CRISPR KO | Wildtype Cas9 or Cas9 nickase is used to cut DNA. When cells attempt to repair DNA breakage, random mutations can be introduced at target sites. | Primarily coding regions | Mainly for functional screens of coding genes |
| CRISPRa | Catalytically inactive Cas9 (dCas9) fused to a transcriptional activator (e.g. VP64) is used to activate genes regulated by target sites. | Usually promoter regions and other noncoding regulatory regions | Gain-of-function screens of coding genes, or screens for regulatory function of noncoding regions |
| CRISPRi | Catalytically inactive Cas9 (dCas9) fused to a transcriptional repressor (e.g. KRAB) is used to inhibit genes regulated by target sites. | Usually promoter regions and other noncoding regulatory regions | Loss-of-function screens of coding genes, or screens for regulatory function of noncoding regions |
Experimental data
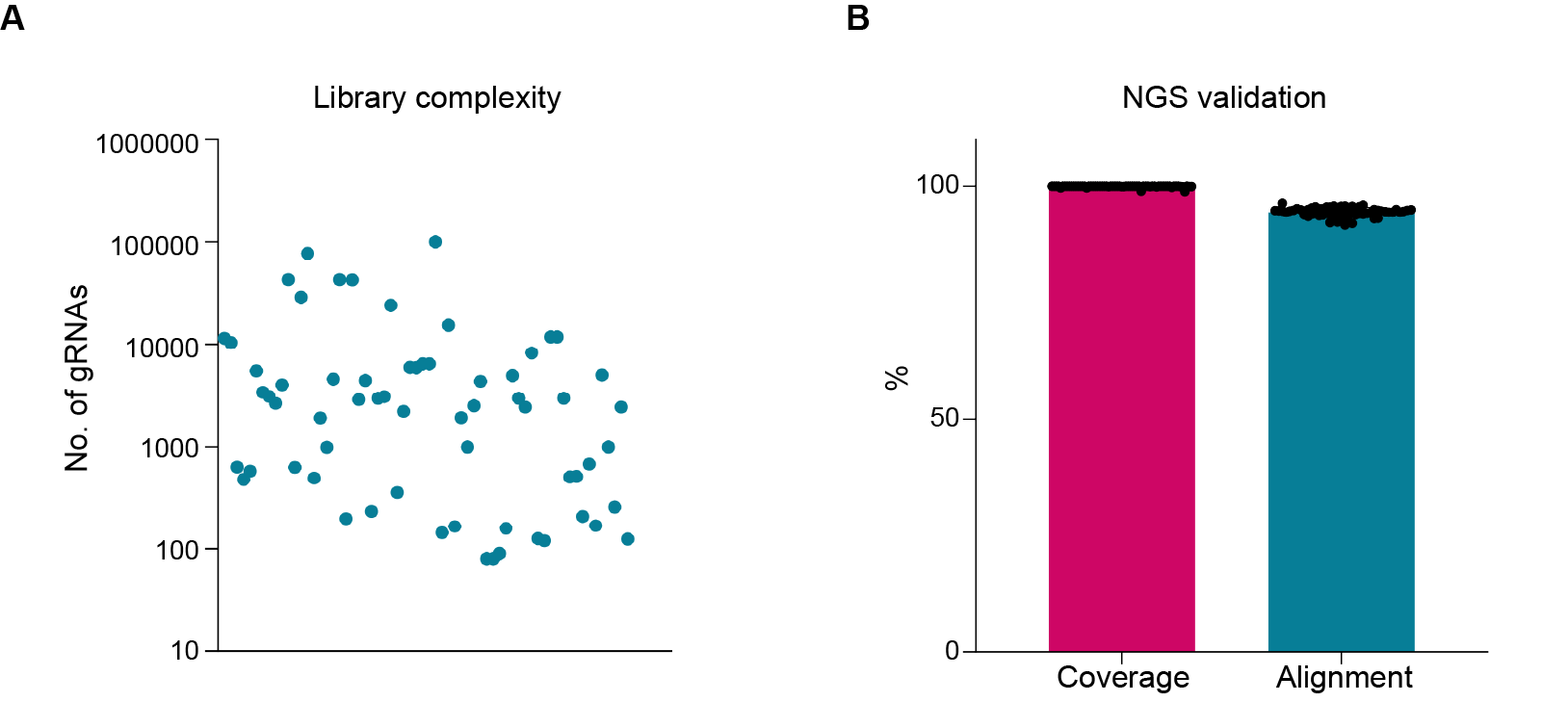
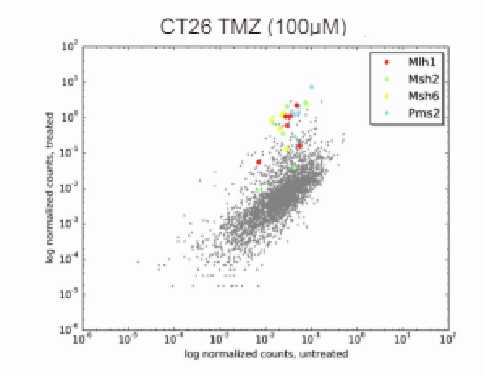
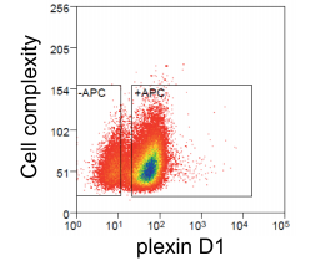
Figure 1. Complexity, coverage, and alignment rate of CRISPR libraries constructed by VectorBuilder. (A) The library complexity ranged from 102 to >105. (B) Both the gRNA coverage and read alignment rate were very high, highlighting the exceptional coverage and low error rate of our libraries.
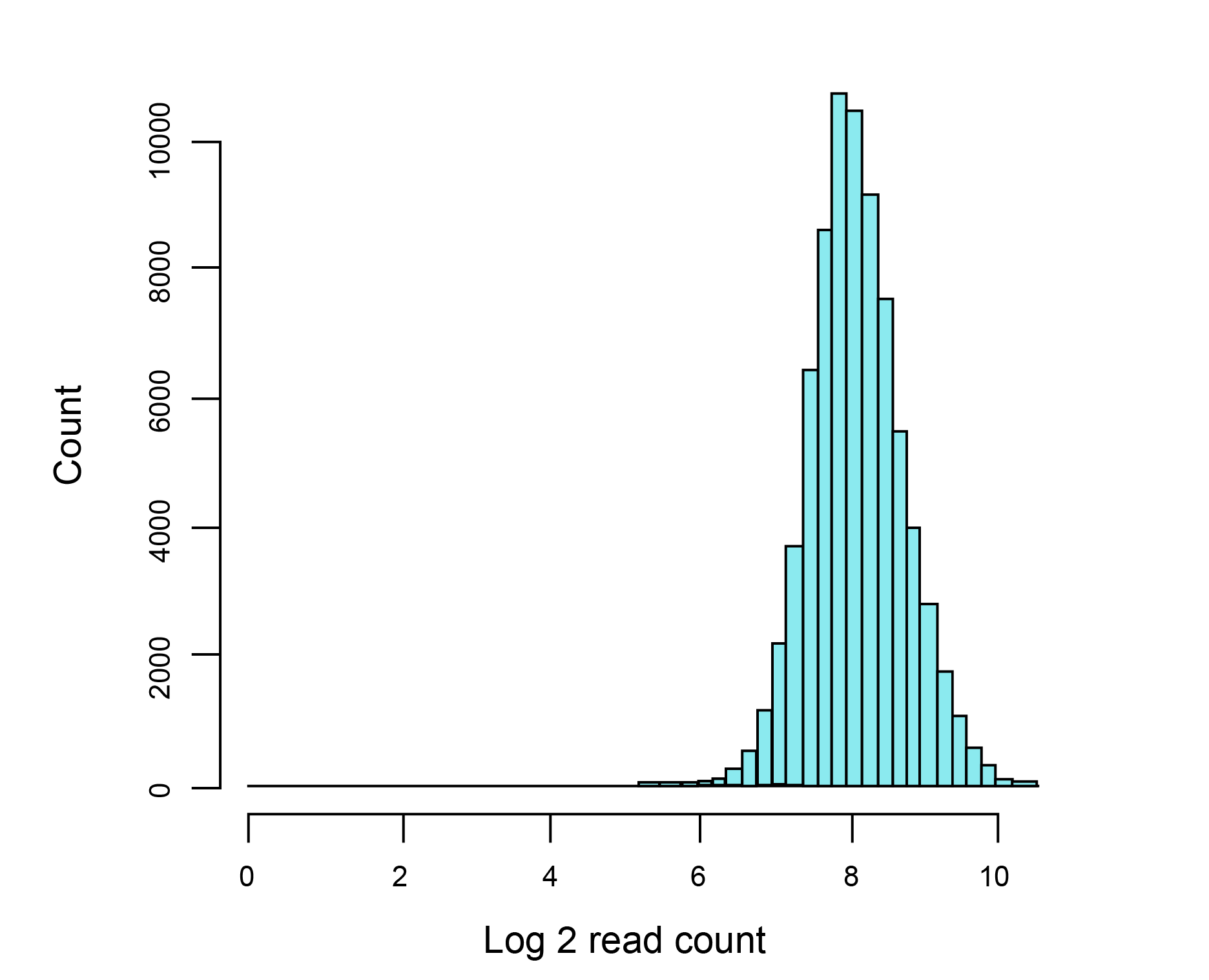
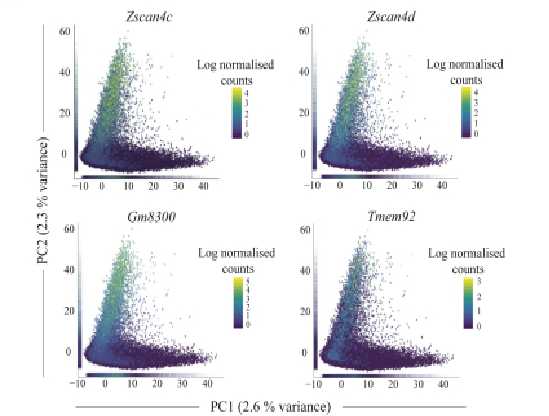
Figure 2. Distribution of gRNA representation in a pooled library consisting of 7.7x104 gRNAs. gRNA read counts were normalized by NGS library size (10 million reads) and plotted in log2 scale. NGS sequencing depth was 300x.
How to Order
Customer-supplied library plasmid pool
If the customer-supplied premade plasmid pools are used, please send us the materials following the Materials Submission Guidelines. Please strictly follow our guidelines to set up shipment to avoid any delay or damage of materials. All customer-supplied materials undergo mandatory QC by VectorBuilder which may incur a $100 surcharge for each item. Please note that production may not be initiated until customer-supplied materials pass QC. For customer-supplied premade plasmid pools, we cannot provide any guarantees regarding the complexity or uniformity of the library.
Resources
FAQ
Dual-gRNA libraries are far more powerful than single-gRNA libraries for knockout screens because the introduction of large deletions by these libraries can have much higher efficiencies in generating loss-of-function mutations. Each CRISPR vector in a dual-gRNA library contains a pair of gRNAs targeting the same gene. When introduced into Cas9-expressing cells, each vector can produce two cuts on the same target gene. Attempts by cells to repair the broken ends of the two cut sites would typically lead to a large deletion spanning the two sites. The two cut sites are designed to flank a functionally important region of the target gene such that a deletion spanning them would most likely lead to the loss of gene function.
For typical knockout screens, a pooled CRISPR library can be constructed in the format of either 1-vector or 2-vector systems. In 1-vector systems, Cas9 or Cas9 variant is co-expressed with gRNA(s) from the same vector, while in 2-vector systems, Cas9 or Cas9 variant and the gRNA(s) are expressed from two separate vectors. Alternatively, in 2-vector systems, gRNA-expressing vectors can be introduced into cell lines that stably express Cas9. The advantages of using the 1-vector system are that it avoids co-transfection/transduction of two different vectors into cells and it is not limited to the availability of a Cas9-expressing stable cell line. However, it is less versatile and can be less efficient in cloning and virus packaging than the 2-vector systems.
VectorBuilder follows the algorithm utilized in CRISPR library design (CLD) to calculate specificity scores for gRNAs. Briefly, for a given gRNA intended to target an N(20)NGG sequence in a species, we search for all potential off-target sites in the genome of that species that have ≤3 mismatches with the target sequence. For each potential off-target site identified this way, a single off-target score is calculated. Scores for all the off-target sites are then used in aggregate to calculate the final specificity score of the gRNA, which is between 0 and 100, with higher values indicating greater targeting specificity.
Please note that specificity score is only a rough guide. Actual targeting efficiency and specificity could depart from what the score predicts. gRNAs with low scores may still work well.