shRNA Gene Knockdown Solutions
VectorBuilder offers a comprehensive collection of shRNA reagents to provide you with the ideal tools for your RNAi experiments. We offer both U6 and miR30-based shRNA systems for providing you with the flexibility to control shRNA expression in different ways based on your experimental needs. We can package all major virus types (e.g. lentivirus, AAV, adenovirus) at various titer scales for delivering your shRNAs into difficult-to-transfect cells. We are also specialized in building pooled shRNA libraries for large-scale loss-of-function screens in mammalian cells. Additionally, our online vector design platform is integrated with shRNA databases for popular species, enabling you to easily select suitable shRNAs to target your gene of interest (GOI).
Highlights
- Highly intuitive online design platform with whole-genome shRNA databases for quick and easy designing of shRNA vectors
- Rich collections of vector backbones and vector components
- Premade and custom-made shRNA libraries available
- 100% sequence validated, fast turnaround and competitive pricing
- Powerful technical support for shRNA selection, vector design and troubleshooting
Offering Details
- Custom shRNA vectors
- Popular shRNA vectors
- shRNA virus
- shRNA (3+1) virus packaging
- Pooled shRNA libraries
- shRNA knockdown stable cell lines
Technical Information
- shRNA-mediated gene knockdown
- Controlling shRNA expression
- Experimental validation
- shRNA databases
Offering Details Price Match
Custom shRNA vectors
Using our high-intuitive online vector design platform, you can choose from over 30 vector backbones (non-viral, viral or transposon) and unlimited combinations of vector components (promoters, fluorescent and drug-selection markers) for expressing your shRNA. Our shRNA databases allow you to easily select shRNAs against your GOI, without the need to design them yourself. We also offer shRNA sensor vectors for testing the knockdown efficiency of the shRNA of interest.
Plasmid DNA preparation and virus packaging can be purchased as downstream services upon finishing the designs of shRNA vectors.
Choose your custom shRNA vectors View more
In addition to the vector systems listed above, we can design and construct IPTG-inducible shRNA expression vectors and Cre-lox based conditional shRNA expression vectors. Just send us a design request!
Popular shRNA vectors
VectorBuilder offers a panel of popular shRNA vectors that express either scramble shRNA or shRNAs targeting popular genes suitable for being used as control vectors in many biological applications. Plasmid DNA preparation and virus packaging can be purchased as downstream services upon adding these vectors into shopping cart.
You can use the vector picker below for ordering U6-based popular shRNA vectors.
For ordering miR30-based popular shRNA vectors, simply send us a design request describing your needs.
shRNA virus
VectorBuilder offers premium quality virus packaging services for lentivirus, AAV and adenovirus in a variety of scales for delivering shRNA into difficult-to-transfect cells. Our proprietary technologies and reagents have greatly improved virus packaging protocols in terms of titer, purity, viability and consistency. Our packaging protocols are also optimized for the viral vector systems used in our vector construction services. As a result, we have a growing base of highly satisfied customers who come back to us again and again for their cloning and virus packaging needs.
Price, turnaround and scales of lentivirus packaging services View more
| Scale | Application | Typical Titer | Minimum Titer | Volume | Price (USD) | Turnaround |
|---|---|---|---|---|---|---|
| Mini | Cell culture | >2x108 TU/ml | >108 TU/ml | 100 ul (4x25 ul) | $199 | 6-12 days |
| Pilot | >4x108 TU/ml | 250 ul (10x25 ul) | $449 | |||
| Medium | >3x108 TU/ml | 1 ml (10x100 ul) | $649 | |||
| Large | >2x109 TU/ml | >109 TU/ml | 1 ml (10x100 ul) | $1,099 | ||
| Ultra-purified medium | Cell culture & in vivo | >2x109 TU/ml | >109 TU/ml | 500 ul (10x50 ul) | $1,399 | |
| Ultra-purified large | 1 ml (10x 100 ul) | $1,699 |
TU = Transduction units (also known as infectious units)
Price, turnaround and scales of AAV packaging servicesView more
| Scale | Application | Typical Titer | Minimum Titer | Volume | Price (USD) | Turnaround |
|---|---|---|---|---|---|---|
| Pilot | Cell culture | >1012 GC/ml | >2x1011 GC/ml | 250 ul (10x25 ul) | $449 | 6-12 days |
| Medium | 1 ml (10x100 ul) | $649 | ||||
| Large | >5x1012 GC/ml | >2x1012 GC/ml | 1 ml (10x100 ul) | $1,099 | ||
| Ultra-purified pilot | Cell culture & in vivo | >2x1013 GC/ml | >1013 GC/ml | 100 ul (4x25 ul) | $1,399 | 7-14 days |
| Ultra-purified medium | 500 ul (10x50 ul) | $1,999 | ||||
| Ultra-purified large | 1 ml (10x100 ul) | $3,099 |
GC = Genome copies
Price, turnaround and scales of adenovirus packaging services View more
| Scale | Application | Typical Titer | Minimum Titer | Volume | Price (USD) | Turnaround |
|---|---|---|---|---|---|---|
| Pilot | Cell culture | >2x1010 IFU/ml | >1010 IFU/ml | 250 ul (10x25 ul) | $649 | 28-35 days |
| Medium | 1 ml (10x100 ul) |
$1,099 |
||||
| Large | >2x1011 IFU/ml | >1011 IFU/ml | 1 ml (10x100 ul) | $1,699 | ||
| Ultra-purified medium | Cell culture & in vivo | >2x1012 VP/ml | >1012 VP/ml | 500 ul (10x50 ul) | $2,099 | |
| Ultra-purified large | 1 ml (10x100 ul) | $2,499 |
IFU = Infectious units; VP = Virus particles
Click to view detailed information on our virus packaging services
shRNA (3+1) virus packaging
It is important to recognize the fact that not all empirically designed shRNAs will work. The potency of an shRNA is determined by several factors including the length of the shRNA, loop structure, GC profile and thermodynamic stability of the shRNA, secondary structure of the target sequence, and off-target matches to other genes. Typically, ~50-70% of shRNAs have a noticeable knockdown effect, and ~20-30% of them have a strong knockdown. Therefore, it is important to test multiple shRNAs to find the most potent one for downstream experiments.
VectorBuilder offers shRNA (3+1) virus packaging services which include 3 custom shRNA viruses targeting your GOI and 1 scramble control virus, enabling you to test multiple shRNAs against your target gene at highly affordable prices. This service is currently available for lentivirus, AAV and adenovirus.
Price, turnaround and scales of shRNA (3+1) virus packaging services View more
| Virus Type | Scale & Deliverable | Application | Price (USD)* | Turnaround** |
|---|---|---|---|---|
| Lentivirus | Pilot | Cell culture | $1,499 | 10-19 days |
| Medium | $1,999 | |||
| Large | $2,999 | |||
| Ultra-purified medium | Cell culture & in vivo | $3,999 | ||
| Ultra-purified large | $4,799 | |||
| AAV | Pilot | Cell culture | $1,499 | 10-19 days |
| Medium | $1,999 | |||
| Large | $2,999 | |||
| Ultra-purified pilot | Cell culture & in vivo | $4,199 | 11-21 days | |
| Ultra-purified medium | $5,699 | |||
| Ultra-purified large | $8,799 | |||
| Adenovirus | Pilot | Cell culture | $2,399 | 34-47 days |
| Medium | $3,599 | |||
| Large | $4,699 | |||
| Ultra-purified medium | Cell culture & in vivo | $6,199 | 34-47 days | |
| Ultra-purified large | $7,499 |
* Price includes the cost of both vector construction and virus packaging.
** Turnaround includes the production time for both vector construction and virus packaging.
Terms and Conditions: VectorBuilder offers a knockdown guarantee for shRNA (3+1) virus packaging services. If none of VectorBuilder's top 3 recommended shRNAs achieve 70% or higher knockdown efficiency for the target gene of interest (GOI), customers can receive store credit rebate equal to 1/3 of the service price. Please note that customers are required to choose the top 3 recommended shRNAs for the guarantee to be applicable.
Use the ordering tool below to order shRNA (3+1) virus packaging for your target genes
Please note that the vector picker above applies for U6-based shRNA vectors only. If you need miR30-based shRNA vectors or if you are not able to find desired shRNAs for your target gene using the vector picker, simply send us a design request describing your needs.
Pooled shRNA libraries
Pooled shRNA libraries can serve as powerful and cost-efficient tools for performing large-scale loss-of-function screens for genes involved in disease pathways, cell responses to drug treatment, developmental processes, gene regulation, etc. We can deliver your library as E. coli stock, plasmid DNA pool, or packaged virus, depending on your needs. Our custom libraries are fully validated by next generation sequencing so that you know exactly what you get.
In addition to custom pooled shRNA library construction, VectorBuilder offers high-quality premade pooled shRNA libraries targeting human and mouse genes. For each species, we provide ready-to-use lentivirus libraries at two scales: Whole Genome (~19,000 RefSeq genes) and Elite Gene (~2,000 most frequently cited genes on PubMed Central). Where possible, each gene is targeted by 5-6 different shRNAs. These libraries have been fully validated by next-generation sequencing and functional assays.
Highlights of our premade shRNA libraries:
- Whole-genome and high-coverage targeting
- Validation of library quality by NGS
- High uniformity
- Available as ready-to-use high-titer lentivirus
- Dual EGFP/Puro marker for efficient and versatile selection or tracking of positively transduced cells
Price and turnaround of premade shRNA librariesView more
| Product Name | No. of Genes | No. of shRNAs | Scale | Catalog No. | Price (USD) | Turnaround | Buy Now |
|---|---|---|---|---|---|---|---|
| Human Elite Gene Pooled shRNA Library | 2,161 | 12,471 |
Medium (>1.0x108 TU/ml, 1 ml) |
LVM(Lib190505-1037bjk) | $1,999 | 7-14 days | |
| Mouse Elite Gene Pooled shRNA Library | 2,233 | 12,472 |
Medium (>1.0x108 TU/ml, 1 ml) |
LVM(Lib190505-1039sgb) | $1,999 | ||
| Human Whole Genome Pooled shRNA Library | 20,593 | 105,233 |
Medium (>1.0x108 TU/ml, 1 ml) |
LVM(Lib230926-1079mym) | $1,999 | ||
|
Plus (>1.0x108 TU/ml, 5 ml) |
LV5M(Lib230926-1079mym) | $2,499 | |||||
| Mouse Whole Genome Pooled shRNA Library | 22,023 | 105,170 |
Medium (>1.0x108 TU/ml, 1 ml) |
LVM(Lib230926-1080rpt) | $1,999 | ||
|
Plus (>1.0x108 TU/ml, 5 ml) |
LV5M(Lib230926-1080rpt) | $2,499 |
* For Elite Gene libraries, the medium scale is sufficient for >40 screens at 100x shRNA coverage. For Whole Genome libraries, the medium scale is sufficient for >5 screens at 100x shRNA coverage, and the plus scale is sufficient for >25 screens at 100x shRNA coverage.
shRNA knockdown stable cell lines
VectorBuilder can custom build shRNA knockdown stable cell lines for applications requiring long-term knockdown of your GOI. To ensure efficient knockdown of your GOI, the top 3 candidate shRNAs based on knockdown score are tested and the one with the best knockdown efficiency is then used for generating the stable cell line via lentivirus transduction. The knockdown level of the cell line is validated by RT-qPCR. Additionally, a series of standard QC assays such as sterility tests and mycoplasma detection are performed for releasing the final cell line products.
Price and turnaround of stable cell line generation servicesView more
| Stable Cell Line Model | Strategy | Deliverable | Price (USD) | Turnaround |
|---|---|---|---|---|
| shRNA Gene Knockdown | Lentivirus-based | Mixed pool (>106 cells/vial, 2 vials) | From $3,999 | 8-13 weeks |
| Two single clones (>106 cells/vial, 2 vials per clone) | From $4,999 | 13-18 weeks |
Technical Information
shRNA-mediated gene knockdown
Short hairpin RNA (shRNAs) are RNA molecules with stem-loop structures that can be used for targeted degradation of mRNA sequences through complementary base-pairing and therefore, are widely used for a variety of RNAi applications. shRNAs can be introduced into target cells using double-stranded DNA vectors, in both viral and non-viral formats. When cells are transfected or transduced with a shRNA vector, the shRNA is transcribed in the nucleus to form a hairpin structure consisting of a sense strand having the same sequence as the mRNA to be silenced, followed by a single-stranded loop and then an antisense strand, which is complementary to the sense strand. The transcribed shRNA exits the nucleus, is processed by Dicer in the cytoplasm and is then loaded onto the RNA-induced silencing complex (RISC) complex for subsequent target mRNA recognition and degradation (Figure 1).
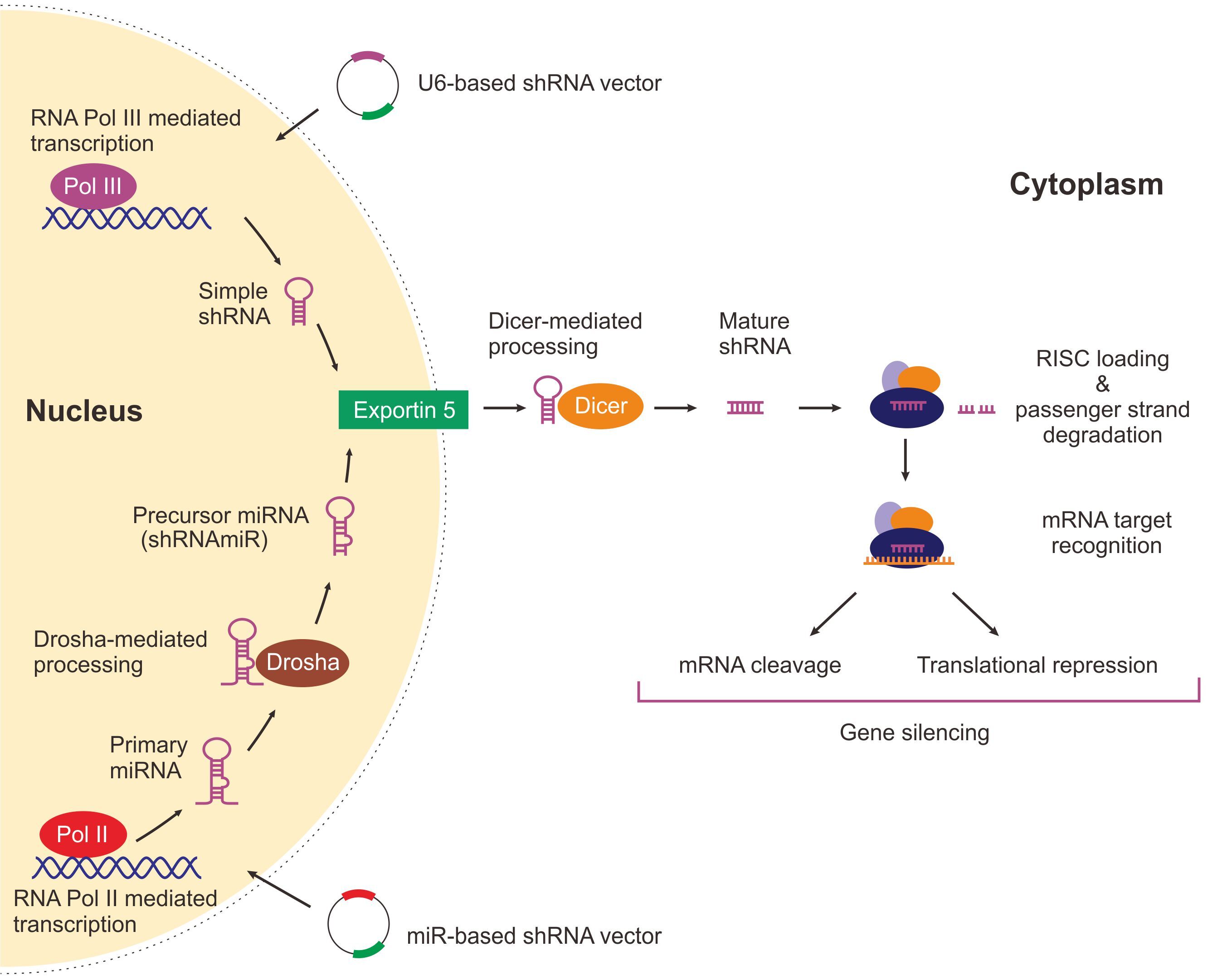
Figure 1 . Mechanisms of U6-based and miR-based shRNA mediated gene expression knockdown.
shRNA-mediated knockdown of gene expression offers several advantages over gene expression knockdown using conventional synthetic small interfering RNAs (siRNAs) and therefore, has gained popularity as the preferred knockdown method for most RNAi applications.
The table below summarizes the advantages of shRNA-mediated gene knockdown over siRNA-mediated gene knockdown:
| shRNA-Mediated Knockdown | siRNA-Mediated Knockdown | |
|---|---|---|
| Delivery method | Transfection or transduction depending on vector type | Transfection |
| Knockdown duration | Long-term | Transient |
| Episomal or stable integration | Can be either episomal or stable depending upon delivery method | Episomal |
| Ability to add selection markers | Yes, fluorescent or drug-selection markers can be added | No |
| Cell-type range | Suitable for a wide range of cell types | Suitable for only cells with high transfection efficiency |
| Off-target effects | Reduced off-target effects | High off-target effects |
| Degradation rate | Low | High |
Controlling shRNA expression
There are two widely used approaches to control shRNA expression on vectors: U6-based shRNA expression and miR-based shRNA expression. While U6-based shRNA vectors drive the expression of simple stem-loop shRNAs transcribed by a RNA Polymerase III promoter such as U6, miR-based shRNA vectors are used for expressing shRNAs adapted with a microRNA scaffold under an RNA Polymerase II promoter. shRNAs expressed by both U6- and miR-based vector systems are processed by similar mechanisms within the cytoplasm ultimately leading to targeted gene silencing. However, after being transcribed within the nucleus, a miR-based shRNA, unlike an U6-based shRNA, is processed in a mechanism similar to that of a primary miRNA due to the presence of endogenous miR-based sequences within its structure (Figure 1).
The presence of RNA polymerase II promoters in miR-based shRNA vectors allows the use of tissue-specific, inducible, or variable-strength promoters, enabling a variety of experimental applications not possible with the constitutive U6 promoter. The ability of RNA polymerase II promoters to efficiently transcribe long transcripts in the miRNA-based shRNA systems provides several additional advantages relative to other knockdown vector systems. Multiple shRNAmiRs can be transcribed as a single polycistron, which is processed to form mature shRNAs within the cell. This allows knockdown of multiple genes or targeting of multiple regions within the same gene using a single transcript. As a result, this vector can be used for expressing either single or multiple shRNAmiRs. Secondly, in this vector system, a user-selected protein coding gene can be positioned within the same polycistron as the shRNAmiRs. The expression of this ORF can be used to directly monitor shRNA transcription (if a marker ORF is used) or can be used for other purposes requiring co-expression of an ORF and shRNA(s).
Despite of the flexibility of controlling shRNA expression using miR-based vectors, we often observed more robust shRNA mediated gene knockdown using U6-based vectors than miR-based vectors. Therefore, unless necessary to use miR-based shRNA vectors, in general we recommend using U6-based shRNA vectors for your gene knockdown experiments.
The table below compares a U6-based shRNA vector system and a miR-based shRNA vector system:
| U6-Based shRNA Vector | miR-Based shRNA Vector | |
|---|---|---|
| shRNA structure | Simple stem-loop shRNA | shRNA adapted with a microRNA scaffold |
| shRNA length | 50-70 nt | >250 nt |
| Promoter | RNA Pol III promoters such as U6 and H1 | RNA Pol II promoters including ubiquitous, tissue-specific and inducible promoters |
| shRNA processing mechanism | Processed by only Dicer in the cytoplasm | Processed by Drosha in the nucleus and Dicer in the cytoplasm |
| No. of shRNAs that can be expressed on one vector | Single shRNA (usually) | Single or multiple shRNAs |
| Ability to express other ORFs in the shRNA transcript | No | Yes |
| Gene knockdown efficiency | Often more robust | Often less robust |
| Toxicity | High cellular toxicity | Decreased cellular toxicity |
Experimental validation
Our lentivirus U6-based shRNA knockdown vector has been validated for highly efficient gene knockdown as shown in Figure 2 below. The comparison between the U6-based and miR30-based shRNA systems is also presented.
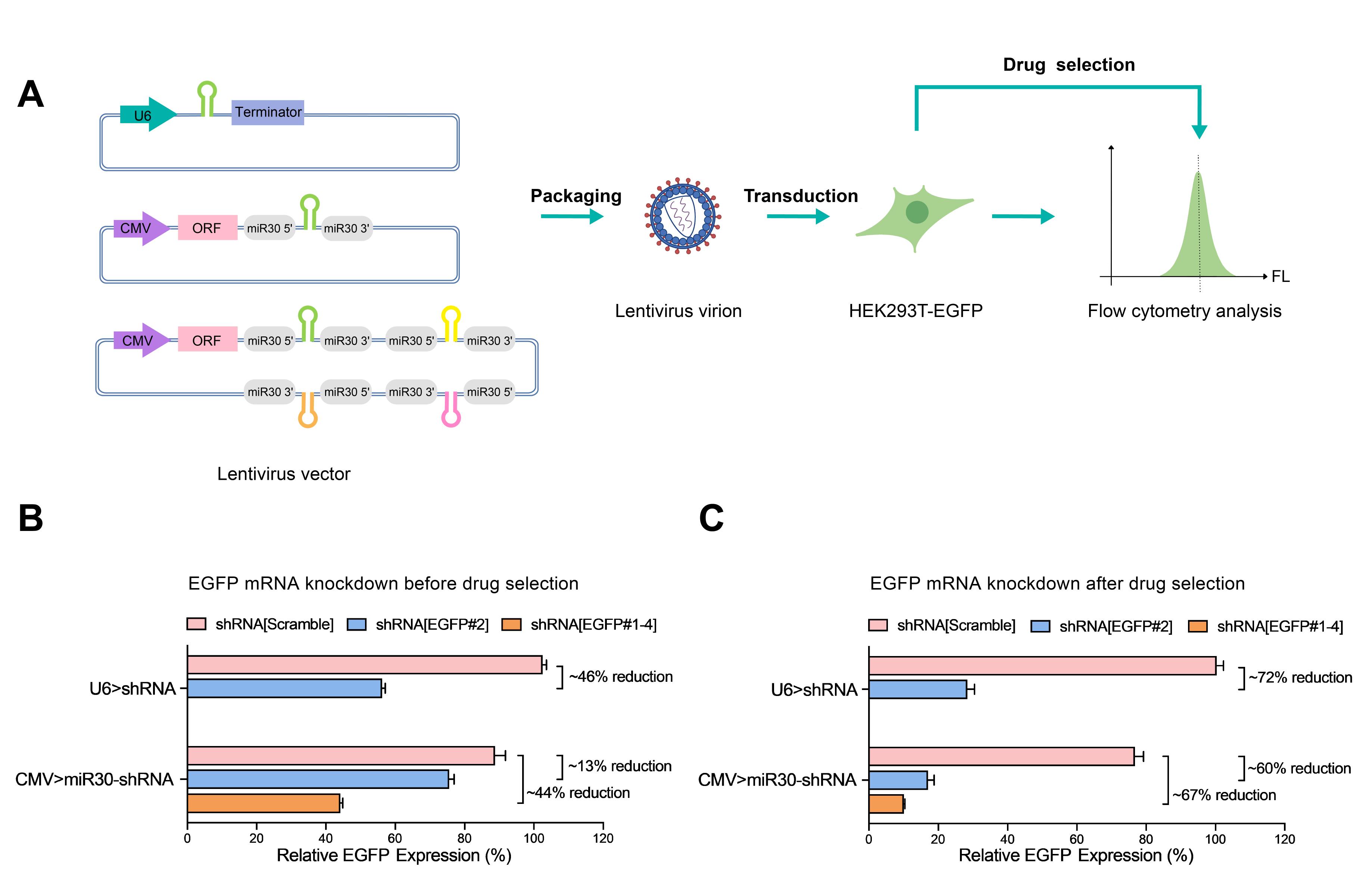
Figure 2. Comparisons of EGFP knockdown through U6-based versus miR30-based shRNA lentiviral systems. (A) Lentiviral vectors carrying the U6-driven shRNA, CMV-driven miR30-based single shRNA, and CMV-driven miR30-based quad shRNA were separately packaged into lentiviral particles. HEK293T cells stably expressing EGFP were transduced with the shRNA lentivirus, and EGFP expression was measured by flow cytometry before and after drug selection using the appropriate antibiotics. (B) Before drug selection, EGFP expression was reduced by ~46% (P<0.001) thru U6-based shRNA, by 13% (P<0.001) thru CMV-driven miR30-based single shRNA, and by 44% (P<0.001) thru CMV-driven miR30-based quad shRNA. (C) After drug selection, EGFP expression was reduced by ~72% (P<0.001) thru U6-based shRNA, by 60% (P<0.001) thru CMV-driven miR30-based single shRNA, and by 67% (P<0.001) thru CMV-driven miR30-based quad shRNA. The relative EGFP expression was calculated by dividing the median fluorescence intensities (MFIs) of the transduced cells by the MFIs of the non-transduced cells. Technical triplicates were performed for the experiment, and SD were presented in the figure. The p-values were calculated based on the Tukey’s test.
shRNA databases
VectorBuilder’s online shRNA vector design tool features optimized shRNA databases for common species, enabling you to design shRNA vectors with high knockdown efficiency for your target genes. For designing shRNAs we apply rules like those used by the RNAi consortium. All scores are ≥0, with mean at ~5, standard deviation at ~5, and 95% of scores ≤15. An shRNA with a knockdown score about 15 is considered to have the best knockdown performance and clonability, while an shRNA with a knockdown score of 0 has the worst knockdown performance or is hard to be cloned.
When you design shRNA vectors on VectorBuilder’s online platform, you will have the option to search for your target genes in our database. Upon entering your gene name, you will see detailed information on all shRNAs against your target gene available in our database, including a link to UCSC Genome Browser to view these shRNAs in the context of genomic sequence and all the transcript isoforms. Our database ranks all available shRNAs for a target gene in order of their decreasing knockdown scores and recommends testing the top 3 shRNAs with the highest knockdown scores. Please note that knockdown scores are only a rough guide. Actual knockdown efficiency could depart significantly from what the scores predict. Target sites with low scores may still work well. Also, please note that targeting 3’ UTR can be as effective as targeting coding region.
VectorBuilder’s online “Resources” contains rich educational materials to help you to successfully plan, execute and troubleshoot your shRNA-based RNAi experiments.
Click to read guides on shRNA vector systemsClick to read guides on various vector components for customizing your shRNA vectors
References
Mol Cell. 9:1327 (2002); Characterization of miR30-based gene knockdown.
Nucleic Acids Res. 34:e53 (2006); Development of miR155-based shRNA vectors.
J Gene Med. 9:620 (2007); Development of IPTG-inducible gene knockdown system.
Proc Natl Acad Sci USA. 101:10380 (2004); Development of Cre-lox-regulated gene knockdown system.
Resources
FAQ
Either shRNA-mediated knockdown or nuclease-mediated knockout (e.g. CRISPR or TALEN) can be valuable experimental approach to study the loss-of-function effects of a gene of interest in cell culture. In order to decide which method is optimal for your specific application, there are a few things you should consider.
Mechanisms
- Knockdown vectors: knockdown vectors express short hairpin RNAs (shRNAs) that repress the function of target mRNAs within the cell by inducing their cleavage and repressing their translation. Therefore, shRNA knockdown vectors are not associated with any DNA level sequence change of the gene of interest.
- Knockout vectors: CRISPR and TALEN both function by directing nucleases to cut specific target sites in the genome. These cuts are then inefficiently repaired by the cellular machinery, resulting in permanent mutations, such as small insertions or deletions, at the sites of repair. A subset of these mutations will result in loss of function of the gene of interest due to frame-shifts, premature stop codons, etc. If two closely positioned cut sites in the genome (i.e. within several kb) are targeted simultaneously, this can also result in the deletion of the intervening region.
Effectiveness
shRNA-mediated knockdown will never completely repress the expression of the target gene. Even for the most effective shRNAs, some residual expression of the target gene will remain. In contrast, in a fraction of treated cells, CRISPR and TALEN can generate permanent mutations which may result in complete loss of gene function.
Consistency and uniformity
shRNA vectors generally provide high cell-to-cell uniformity within the pool of treated cells and very consistent results between experiments. In contrast, CRISPR and TALEN produce results that are highly non-uniform from cell to cell due to the stochastic nature of the mutations introduced. To fully knock out the gene of interest in a cell, all copies of the gene in the cell must be knocked out. Given that normal cells have two copies of any gene (except for X- or Y-linked genes) while cancer cells can have more than two copies, such full knockout cells may represent a very small fraction of all the treated cells. For this reason, nuclease-mediated knockout experiments require the screening of clones by sequencing to identify the subset in which all copies of the gene of interest have been knocked out.
Off-target effects
Off-target effects have been reported for both shRNA-mediated knockdown and nuclease-mediated knockout. The off-target phenotype(s) can be estimated by using multiple different shRNAs to target the same gene. If a gene knocked down by multiple different shRNAs results in consistent phenotype(s), then it argues against the phenotype(s) being caused by off-target effects. For CRISPR- or TALEN-mediated knockout, multiple clones containing loss-of-function mutations should be analyzed in order to account for any phenotype(s) that may be due to off-target mutations. Additionally, bioinformatically identified off-target sites could be sequenced in the clones to see if they have been mutated.
Design your homologous recombination donor vector onlineNot all shRNAs will work
Based on our experience and feedback from our customers, we know that generally when 3 or 4 shRNAs are tested for any arbitrary gene, typically 2 or 3 produce reasonable to good knockdown. However, when using shRNAs, it is important to recognize the fact that not all shRNAs will work. Typically, ~50-70% of shRNAs have noticeable knockdown effect, and ~20-30% of them have strong knockdown. If you try a few shRNAs targeting a specific gene, it is possible that by chance, none will produce satisfactory knockdown. When this happens, the best approach is to try more shRNAs, especially the ones that have literature validation. Many researchers also use a “cocktail” of shRNAs (i.e. mixture of different shRNAs) targeting the same gene, which sometimes can improve knockdown efficiency.
The assay for validating the knockdown of your gene is not performed properly
The most common and sensitive assay to evaluate shRNA knockdown efficiency is RT-qPCR. Sometimes, you may need to try several pairs of primers, and then choose the most specific and efficient pair to use. In general, the RT-qPCR primers should span exon-exon junction if possible to avoid amplifying genomic DNA. When using a new pair of primers, we recommend that you run the PCR product on an agarose gel to verify the band, or even validate the PCR product by sequencing. You should always include minus-RT control in RT-qPCR to better estimate the level of genomic DNA contamination. You can use NCBI primer designing tool to help you better examine the quality of your primers in silico.
Knockdown efficiency can also be assessed by Western blot. However, Western blot is notoriously prone to false positive bands from non-specific antibody binding, which could mistakenly lead to the interpretation that there is no knockdown. Care must therefore be taken to make sure that the antibody used is indeed specific to the gene of interest.
The shRNA might only target a subset of transcript isoforms of your gene
When designing shRNA, we generally recommend those that can target as many transcript isoforms of the gene as possible, unless you are only interested in knocking down a particular isoform. VectorBuilder has created shRNA databases that contain optimized shRNAs for common species. If you design shRNA vectors on VectorBuilder, when you insert the shRNA component into the vector, you will have the option to search the target gene in our database. Then, you will see the detailed information of all the available shRNAs we designed for you, including a link to UCSC Genome Browser to view these shRNAs in the context of genomic sequence and all the transcript isoforms.
VectorBuilder applies rules similar to that used by the RNAi consortium (TRC) to design and score shRNAs. For each given RefSeq transcript, we search for all possible 21mers that are considered as candidate target sites. Candidates are excluded if they contain features thought to reduce knockdown efficiency/specificity or cloneability, including a run of ≥4 of the same base, a run of ≥7 G or C, GC content <25% or >60%, and AA at the 5’ end. Knockdown scores are penalized for candidates that contain internal stem-loop, high GC content toward the 3’ end, known miRNA seed sequences, or off-target matches to other genes. For genes with alternative transcripts, target sites that exist in all transcripts are given higher scores.
All scores are ≥0, with mean at ~5, standard deviation at ~5, and 95% of scores ≤15. An shRNA with a knockdown score about 15 is considered to have the best knockdown performance and cloneability, while an shRNA with a knockdown score of 0 has the worst knockdown performance or is hard to be cloned.
Please note that knockdown scores are only a rough guide. Actual knockdown efficiency could depart significantly from what the scores predict. Target sites with low scores may still work well. Also, please note that targeting 3’ UTR can be as effective as targeting coding region.




