Therapeutic IVT RNA
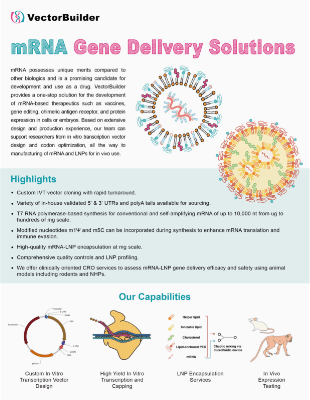
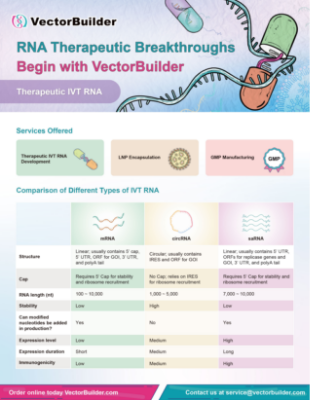
VectorBuilder offers a comprehensive platform for the development of RNA therapeutics including vaccines, gene replacement, chimeric antigen receptor (CAR), and gene editing. Leveraging our extensive design and production experience with premade and custom RNAs, our team can support in vitro transcription (IVT) RNA synthesis and lipid nanoparticle (LNP) encapsulation for a variety of RNA-based expression systems. Researchers can accelerate the development of their RNA therapeutics with our CRO services, offering in vitro and in vivo candidate screening and functional testing.
Highlights
 Comprehensive Platform Offering research and clinical grades of RNA, a variety of RNA systems, customizable IVT RNA production and LNP encapsulation, and a full suite of CRO and CDMO services
Comprehensive Platform Offering research and clinical grades of RNA, a variety of RNA systems, customizable IVT RNA production and LNP encapsulation, and a full suite of CRO and CDMO services Experts in RNA Development Our team comprises scientists with expertise of the full scope of considerations for therapeutic IVT vector design, sequence optimization and screening, and process development
Experts in RNA Development Our team comprises scientists with expertise of the full scope of considerations for therapeutic IVT vector design, sequence optimization and screening, and process development Highest Quality and Consistency State-of-the-art facilities and equipment providing the highest purity and consistency, ensured by a full panel of quality control assays
Highest Quality and Consistency State-of-the-art facilities and equipment providing the highest purity and consistency, ensured by a full panel of quality control assaysServices Offered
Products Offered
Documents
Brochures & Flyers User InstructionsCertificate of Analysis (COA)
Material Safety Data Sheet (MSDS)