Dual-gRNA CRISPR Libraries
VectorBuilder offers high-quality, premade dual-gRNA lentivirus libraries for CRISPR-based whole-genome knockout screens in human and mouse cells. Wherever possible, each gene is targeted redundantly by 4-6 different gRNA pairs in separate vectors. These libraries can serve as powerful and highly cost-effective tools for performing genome-wide loss-of-function screens. Additionally, we can design and construct custom pooled gRNA libraries for your target gene list.
Types of dual-gRNA CRISPR libraries offered
- Ready-to-use, pooled, lentivirus libraries targeting the entire human genome (20,048 human genes)
- Ready-to-use, pooled, lentivirus libraries targeting the entire mouse genome (20,493 mouse genes)
- Custom pooled gRNA libraries (available as E.coli stock, DNA or recombinant virus)
Ordering Information Price Match
| Product Name | No. of Genes | No. of gRNA pairs | Scale | Catalog No. | Price (USD) | Turnaround | Buy Now |
|---|---|---|---|---|---|---|---|
| Human Whole-Genome Dual-gRNA Library | 20,048 | 91,926 |
Medium (>1.0x108 TU/ml, 1 ml) |
LVM(Lib190505-1046fgb) | $2,999 | 7-14 days | |
|
Plus (>1.0x108 TU/ml, 5 ml) |
LV5M(Lib190505-1046fgb) | $3,499 | |||||
| Mouse Whole-Genome Dual-gRNA Library | 20,493 | 90,344 |
Medium (>1.0x108 TU/ml, 1 ml) |
LVM(Lib190505-1050kpm) | $2,999 | ||
|
Plus (>1.0x108 TU/ml, 5 ml) |
LV5M(Lib190505-1050kpm) | $3,499 |
Deconvolution service
Not sure how to identify gRNA hits by NGS after conducting your CRISPR library screen? You can simply send the genomic DNA of your samples to VectorBuilder, and we will prepare the NGS libraries, perform high-throughput sequencing, analyze the data, and deliver the accurate counts of gRNAs in each sample to you in just a few weeks.
For deconvolution service, please Request Design Support from us.
Technical Information
Product highlights View more
Unique dual-gRNA lentiviral vector design
As the only commercially available whole genome dual-gRNA lentivirus libraries, each lentiviral vector in the libraries contains a pair of gRNA expression cassettes and an EGFP/puromycin dual marker cassette (Figure 1). The paired gRNAs from the same vector are expressed simultaneously when the vector is introduced into cells, targeting two separate sites on the same gene, resulting in two CRISPR cut sites that then lead to large loss-of-function deletions for the knockout screen. The two gRNAs in each vector are coupled with two different U6 promoters (human U6 and macaque U6) and two different gRNA scaffolds (described in Nat Methods. 14:573 (2017)) that are distinct in sequence but equivalent in function. This reduces unwanted recombination between the two gRNA cassettes and also allows the PCR amplification and sequencing by NGS of either the upstream or the downstream gRNA, or both, from cells transduced with the libraries.
Figure 1. Map of the dual-gRNA library vector.
Click to view fully annotated map and sequence of the dual gRNA library vector
Whole genome and high-coverage targeting
The human and mouse dual-gRNA lentivirus libraries target 20,048 human genes and 20,493 mouse genes, respectively. On average, each gene is targeted by 4-6 different gRNA pairs selected by a set of parameters, including 1) off-target scores to minimize off-target effect; 2) locations of the cut sites to ensure that deletions spanning the two sites would most likely lead to the loss of gene function; 3) distances between the cut sites to bias the resulting mutations toward large deletions across the cut sites rather than local mutations at each cut site, and 4) number of alternative transcripts affected in each gene to maximize the number of transcripts being knocked out. All the gRNA cut sites are located within annotated exons. Therefore, even if large deletions fail to be created between sites, any local mutations within a cut site (typically small deletions) can also generate mutations that are likely to disrupt gene function. Detailed lists of target genes and gRNA can be found under “Documents”.
Validation of library quality by NGS
The dual-gRNA lentivirus CRISPR libraries have been validated by NGS in which both gRNAs of the pairs have been fully sequenced. This showed that >85% and >75% of the total sequencing reads matched the designed gRNA pairs for human and mouse libraries, respectively. Furthermore >99% and >97% of the designed gRNA pairs for human and mouse libraries were detected by NGS, respectively. Thus, both libraries have excellent coverage of the designed gRNA pairs with low error rate. To our knowledge, these are the highest-quality dual-gRNA libraries that have been reported.
High uniformity
The representation of gRNA pairs in both libraries are highly uniform (Figure 2).
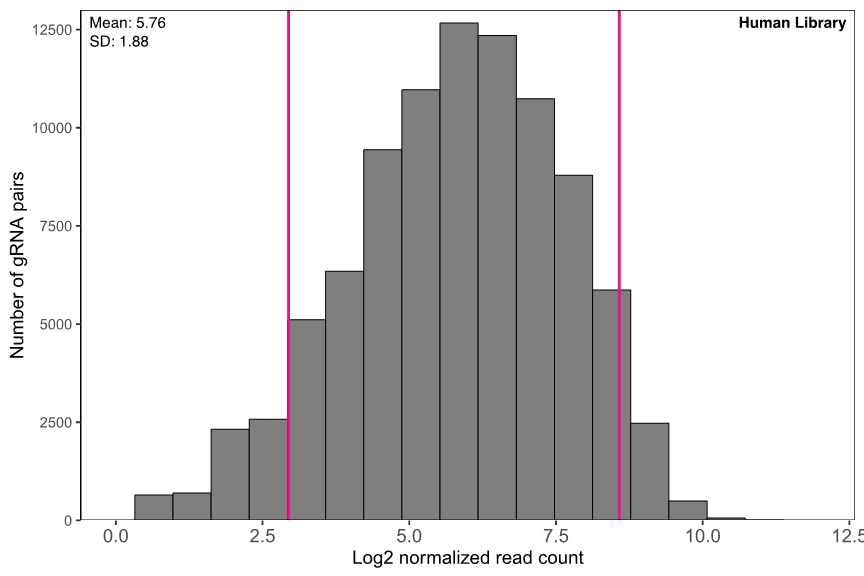
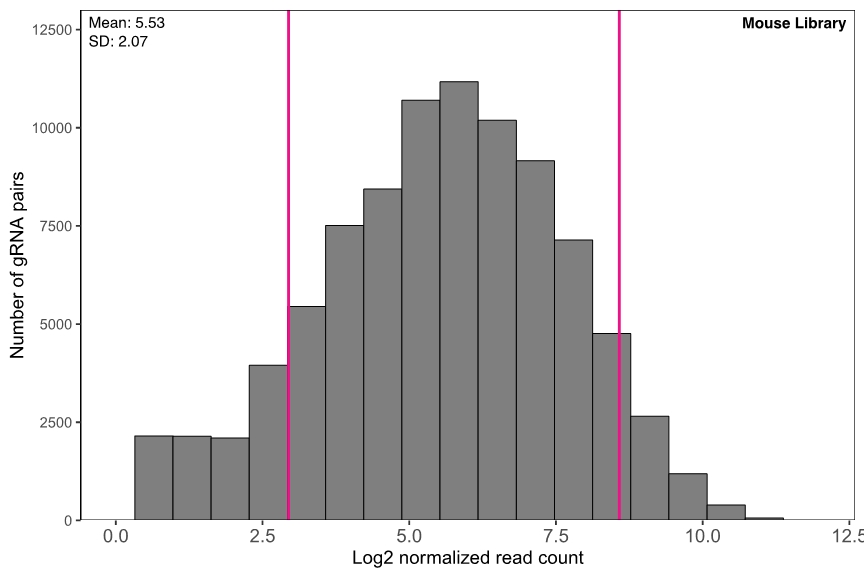
Figure 2. Representation of gRNA pairs in different pooled libraries. gRNA read counts are normalized by NGS library size (10 million reads) and plotted in log2 scale.
Well-established lentiviral system and ready-to-use high-titer lentivirus
The pooled CRISPR libraries are expressed in the third-generation lentiviral vector system, which is a highly efficient expression system in a wide variety of cells. This lentiviral vector system is ideal for in vitro genetic screens since it can introduce gRNAs into cells permanently, efficiently and relatively uniformly. All pooled CRISPR libraries are provided as ready-to-use lentivirus with high functional titer (>108 TU/ml), which saves you time in virus packaging and titer measurement. The third-generation lentiviral vector system is optimized for improved biosafety given its incompetent self-replication feature.
Dual marker expression for efficient and versatile selection or tracking of positively transduced cells
A dual-marker expression cassette of EGFP and puromycin resistance gene (Puro) is expressed from the lentiviral vector, allowing for selection of positively transduced cells by puromycin and visual tracking by green fluorescence (Figure 3).
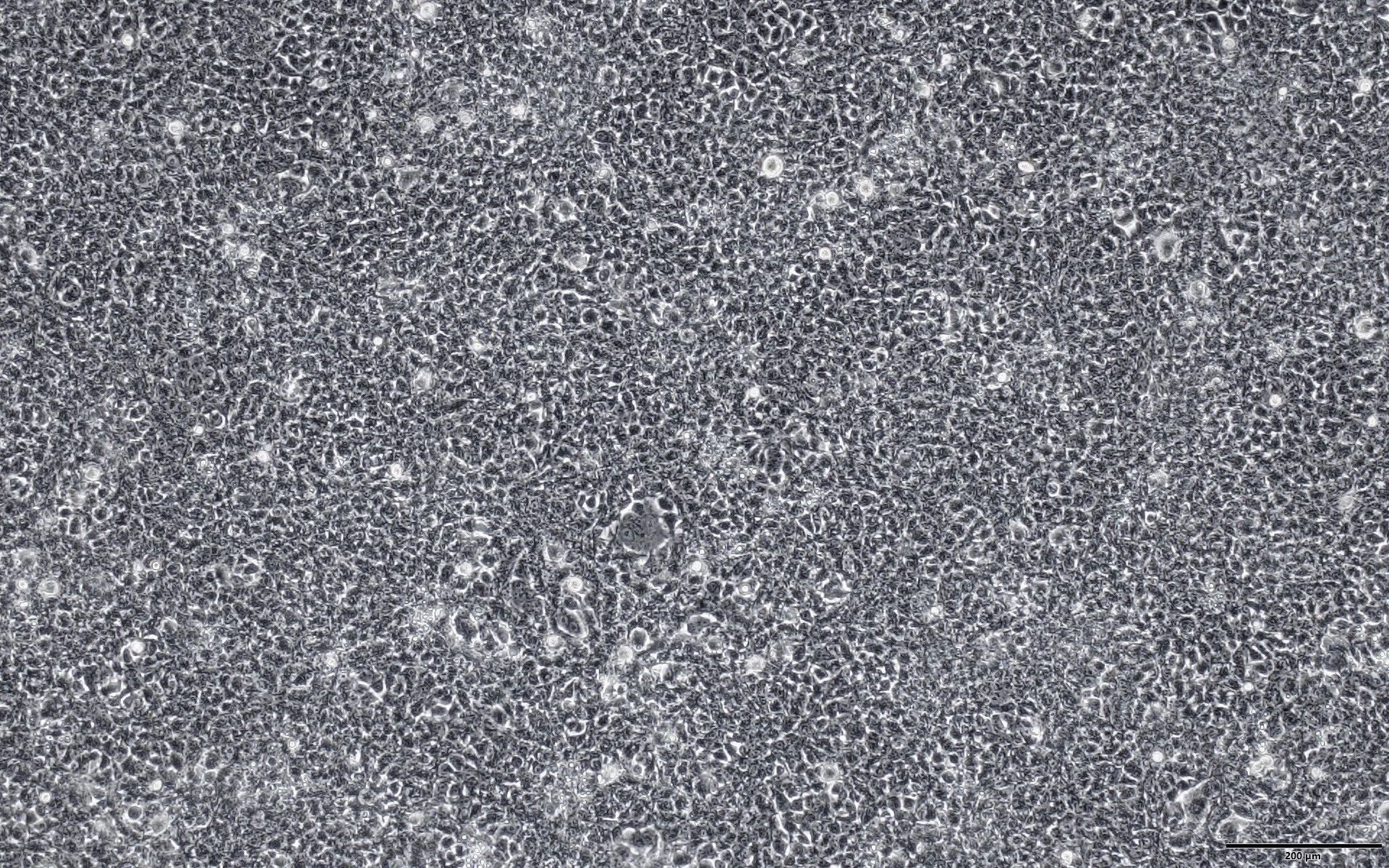
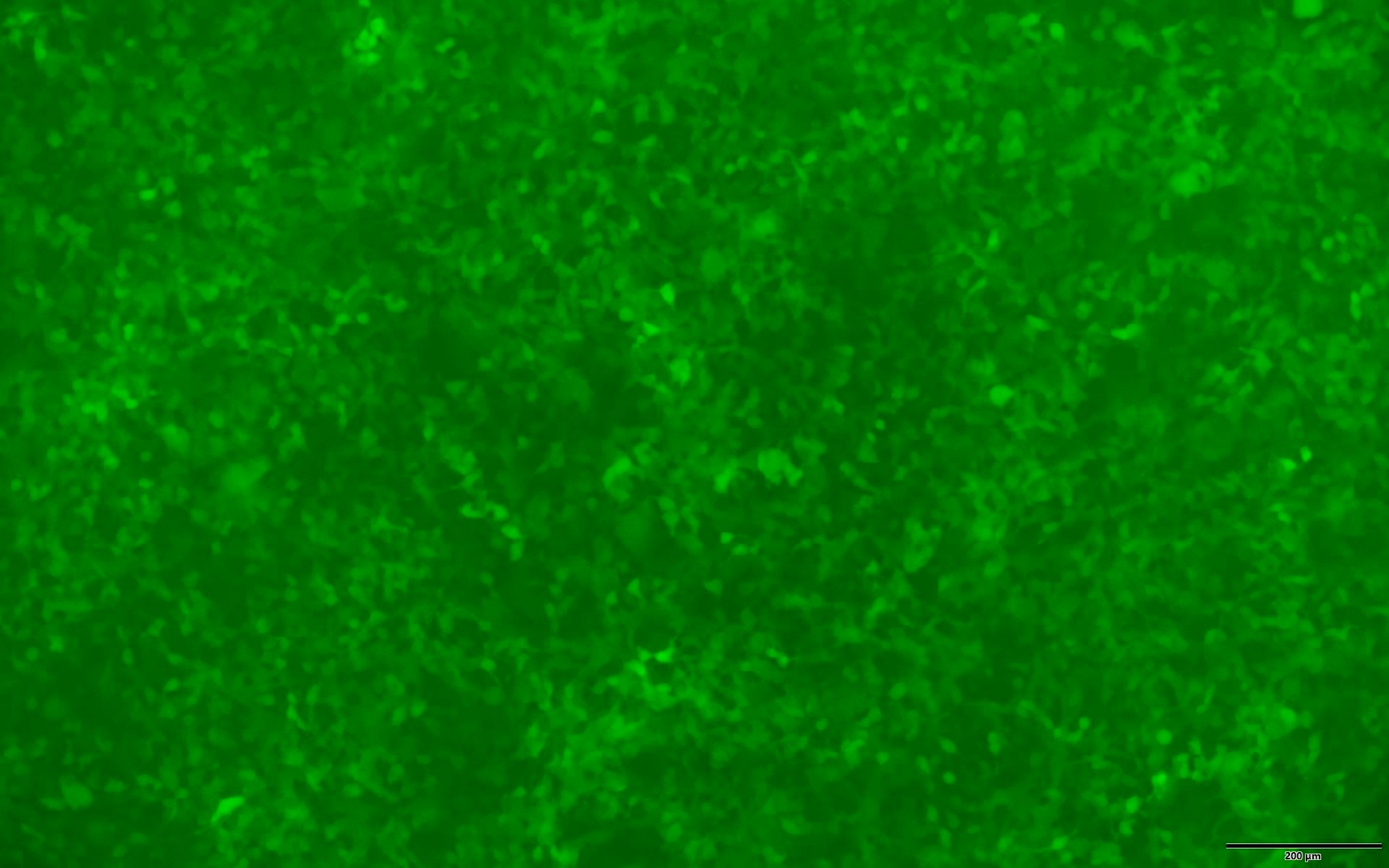
Figure 3. EGFP expression in 293T cells transduced with Human Whole Genome Dual-gRNA Lentivirus Library (MOI=10) after 4 days of puromycin selection (1.5 ug/ml). Left: bright field. Right: EGFP. Scale bars, 200 um.
Workflow of pooled CRISPR library-mediated genetic screen View more
A typical workflow for CRISPR/Cas9 screens using pooled dual-gRNA lentivirus libraries is shown in Figure 4. First, Cas9 expressing cells of interest are transduced with the lentivirus library and positively transduced cells are selected by the marker gene(s) carried on the lentiviral vector (e.g. drug-selection or fluorescence marker). Next, positively transduced cells are split into a reference population and an experimental population. Then, the experimental population is subjected to particular selective pressure (e.g. drug treatment or repeated passaging) to identify cells with the phenotype of interest. There are three major types of screening strategies: 1) viability screens that search for gRNAs enriched or depleted in surviving cells when exposed to the selective pressure; 2) reporter screens that look for gRNAs enriched in cells associated with either high or low reporter expression ( e.g. gRNAs targeting transcription factors that modulate reporter gene expression); 3) behavior screens that identify gRNAs affecting genes associated with cell invasion, migration, etc. After screening, both experimental and reference cells are harvested. Enriched or depleted gRNAs in the experimental group relative to the reference group are identified using Sanger sequencing or next-generation sequencing (NGS). Candidate genes that are theoretically targeted by the enriched or depleted gRNAs can be further investigated with downstream functional studies.
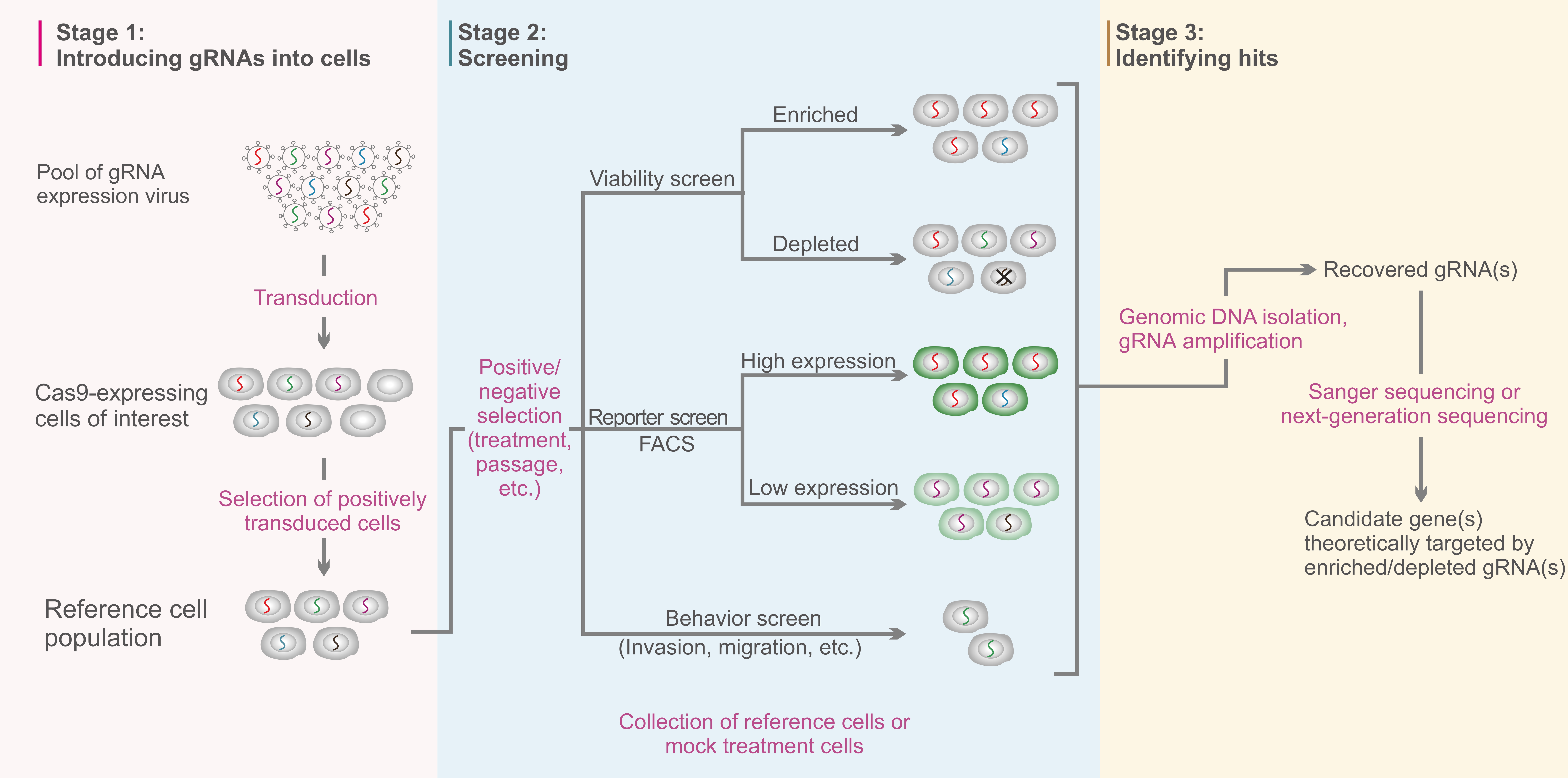
Figure 4. Workflow of CRISPR-based knockout screens employing pooled dual-gRNA lentivirus libraries. Adapted from Acta Biochim Biophys Sin 44:103-112 (2012).
Documents View more
Full Lists of Target Genes and gRNA Pairs
Brochures & Flyers
FAQ
What are the advantages of dual-gRNA libraries compared to single-gRNA libraries?
Dual-gRNA libraries are far more powerful than single-gRNA libraries for knockout screens because the introduction of large deletions by these libraries can have much higher efficiencies in generating loss-of-function mutations. Each CRISPR vector in a dual-gRNA library contains a pair of gRNAs targeting the same gene. When introduced into Cas9-expressing cells, each vector can produce two cuts on the same target gene. Attempts by cells to repair the broken ends of the two cut sites would typically lead to a large deletion spanning the two sites. The two cut sites are designed to flank a functionally important region of the target gene such that a deletion spanning them would most likely lead to the loss of gene function.
For CRISPR knockout screens, should I use 1-vector systems or 2-vector systems?
For typical knockout screens, a pooled CRISPR library can be constructed in the format of either 1-vector or 2-vector systems. In 1-vector systems, Cas9 or Cas9 variant is co-expressed with gRNA(s) from the same vector, while in 2-vector systems, Cas9 or Cas9 variant and the gRNA(s) are expressed from two separate vectors. Alternatively, in 2-vector systems, gRNA-expressing vectors can be introduced into cell lines that stably express Cas9. The advantages of using the 1-vector systems are that it avoids co-transfection/transduction of two different vectors into cells and it is not limited to the availability of a Cas9-expressing stable cell line. However, it is less versatile and can be less efficient in cloning and virus packaging than the 2-vector systems.
How is gRNA specificity score calculated?
VectorBuilder follows the algorithm utilized in CRISPR library design (CLD) to calculate specificity scores for gRNAs. Briefly, for a given gRNA intended to target a N(20)NGG sequence in a species, we search for all potential off-target sites in the genome of that species that have ≤3 mismatches with the target sequence. For each potential off-target site identified this way, a single off-target score is calculated. Scores for all the off-target sites are then used in aggregate to calculate the final specificity score of the gRNA, which is between 0 and 100, with higher values indicating greater targeting specificity.
Please note that specificity scores are only a rough guide. Actual targeting efficiency and specificity could depart from what the scores predict. gRNAs with low scores may still work well.
Related Services
Library constructionCRISPR genome editing solutions
Vector cloning
Virus packaging



