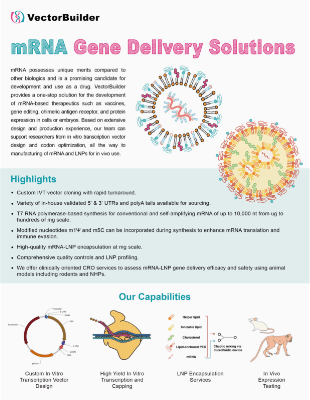
Therapeutic IVT RNA Development
VectorBuilder offers comprehensive CRO services for the development of RNA therapeutics including vaccines, gene replacement, chimeric antigen receptor (CAR), and gene editing. Our team of experts understand key considerations for therapeutic design and process development, and can utilize their know-how to enhance the efficacy, safety, and manufacturability of RNA drugs. For large-scale manufacturing of RNA therapeutics, check out our CDMO services.
Talk to Our Experts




IVT Vector
Design & Cloning
Design & Cloning
IVT RNA Production
LNP Encapsulation
Quality Control (QC)
Functional Validation
IVT Vector Design & Cloning
- Royalty-free IVT backbone with no IP constraints for commercial use
- Proprietary sequence optimization for optimal expression
- Robust cloning and transcription of 120 nt (and longer) polyA tail
- UTR optimization
- Coding sequence optimization
- PolyA integrity
- PolyA length and GOI expression
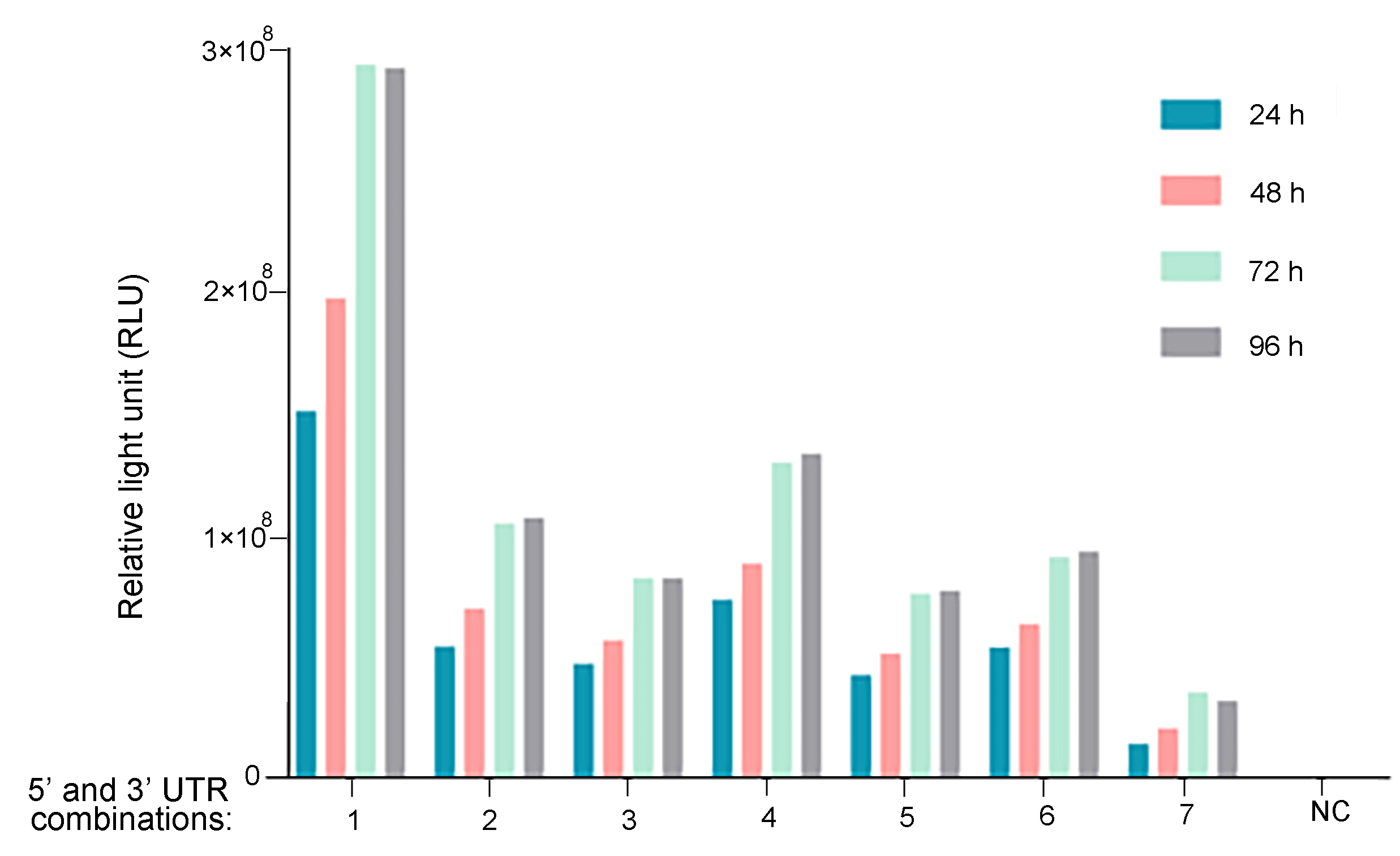
Figure 1. UTR optimization for improved mRNA expression. Different 5' and 3' UTR combinations were tested regulating Gaussia luciferase expression in vitro. HEK293T cells were seeded on 12-well plates at a density of 2.3x105 cells per well. Cells were transfected with 1 ug of mRNA per well. At 6 h, 24 h, 48 h, 72 h, and 96 h post-transfection, Gaussia luciferase activities were measured from cell culture medium.
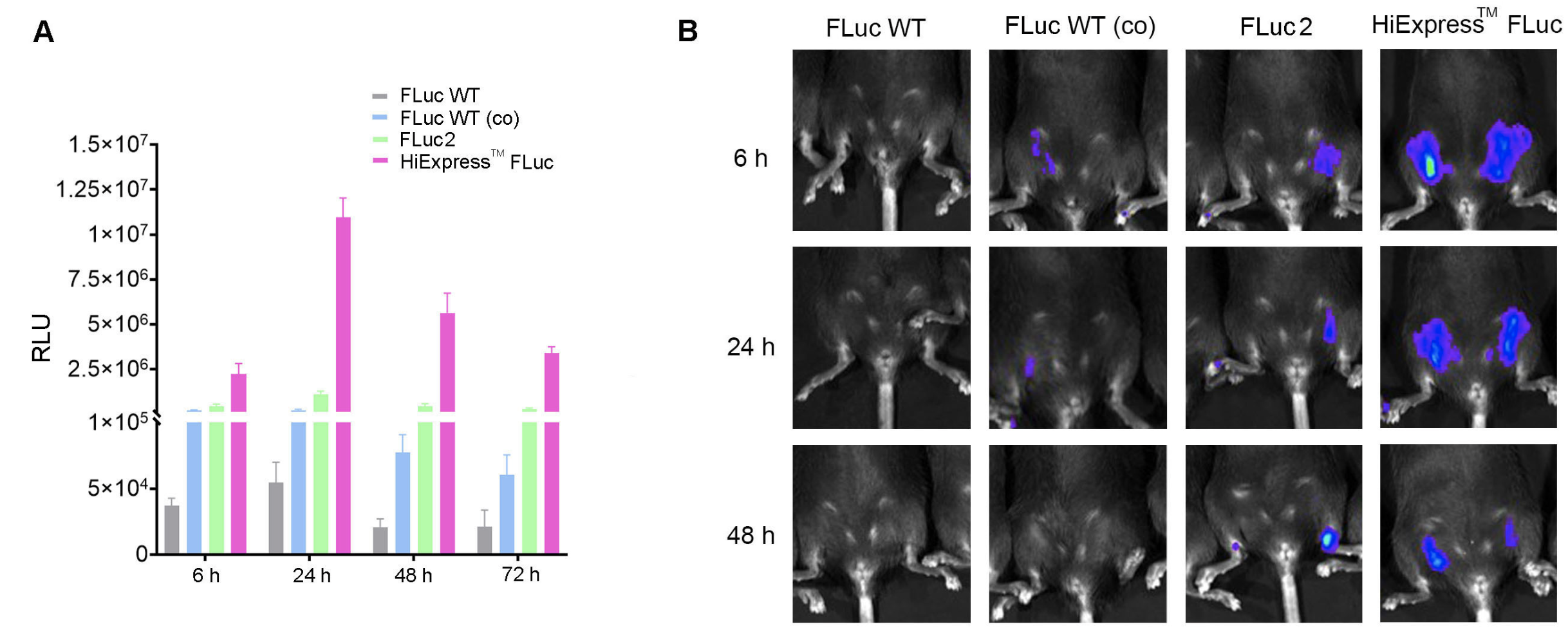
Figure 2. Codon optimization increases mRNA expression in vitro and in vivo. (A) Expression of HiExpress™ Firefly Luciferase mRNA and other luciferase mRNA in HEK293T cells. Cells grown on a 12-well plate were transfected with 0.5 ug of mRNA per well and luciferase activity was measured at 6 h, 24 h, 48 h, and 72 h post-transfection. (B) Luciferase activity measured in adult C57BL/6 mice injected intramuscularly with 30 ug of LNP-mRNA at 6 h, 24 h, and 48 h post-injection. FLuc WT indicates wild-type firefly luciferase. FLuc WT (co) indicates codon-optimized wild-type firefly luciferase. FLuc2 indicates Luc2 firefly luciferase.
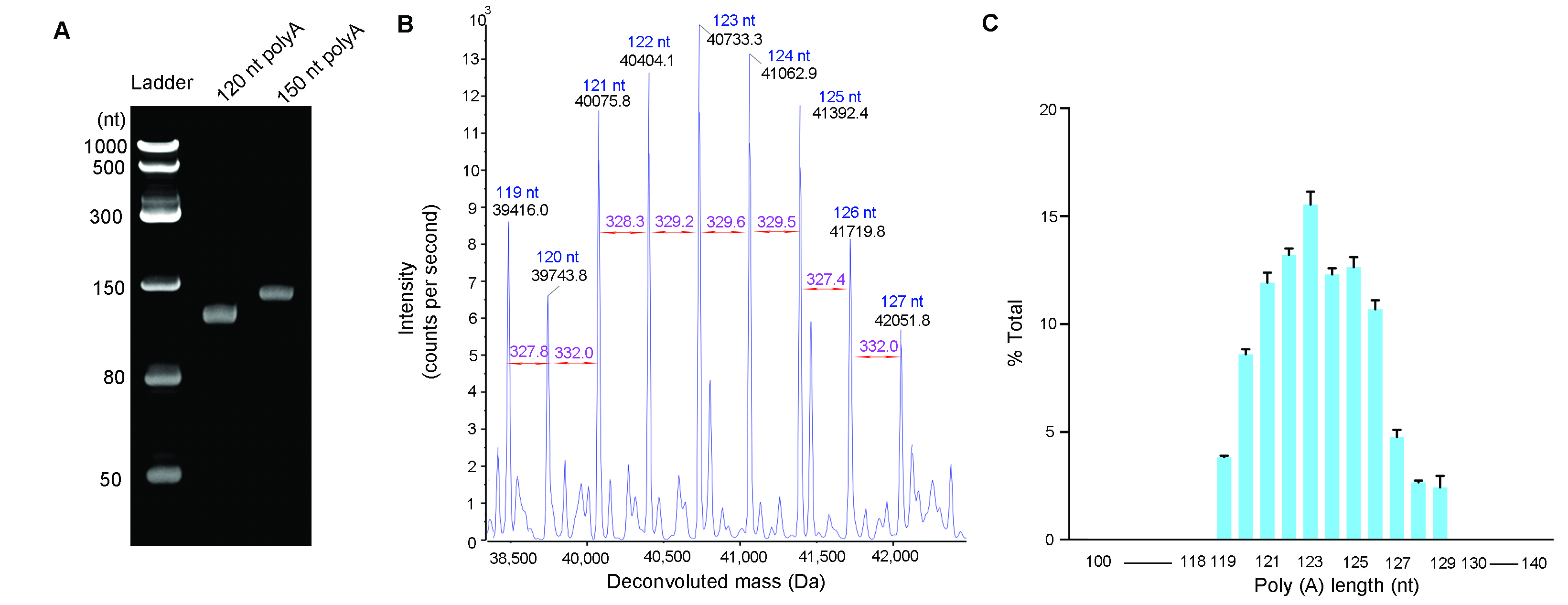
Figure 3. PolyA tail size analysis. PolyA tails were cleaved from IVT mRNA using ribonuclease T1 and isolated by oligo dT affinity chromatography. (A) Isolated polyA tails analyzed by Urea-PAGE gel electrophoresis. (B) Isolated polyA tails analyzed by LC-MS. Deconvoluted spectrum was generated from 120 pmol of polyA tails with an expected size of 120 nt. (C) Size distribution of polyA tails with an expected size of 120 nt. Error bars represent standard deviation from triplicates. Weighted average length is 123 nt.
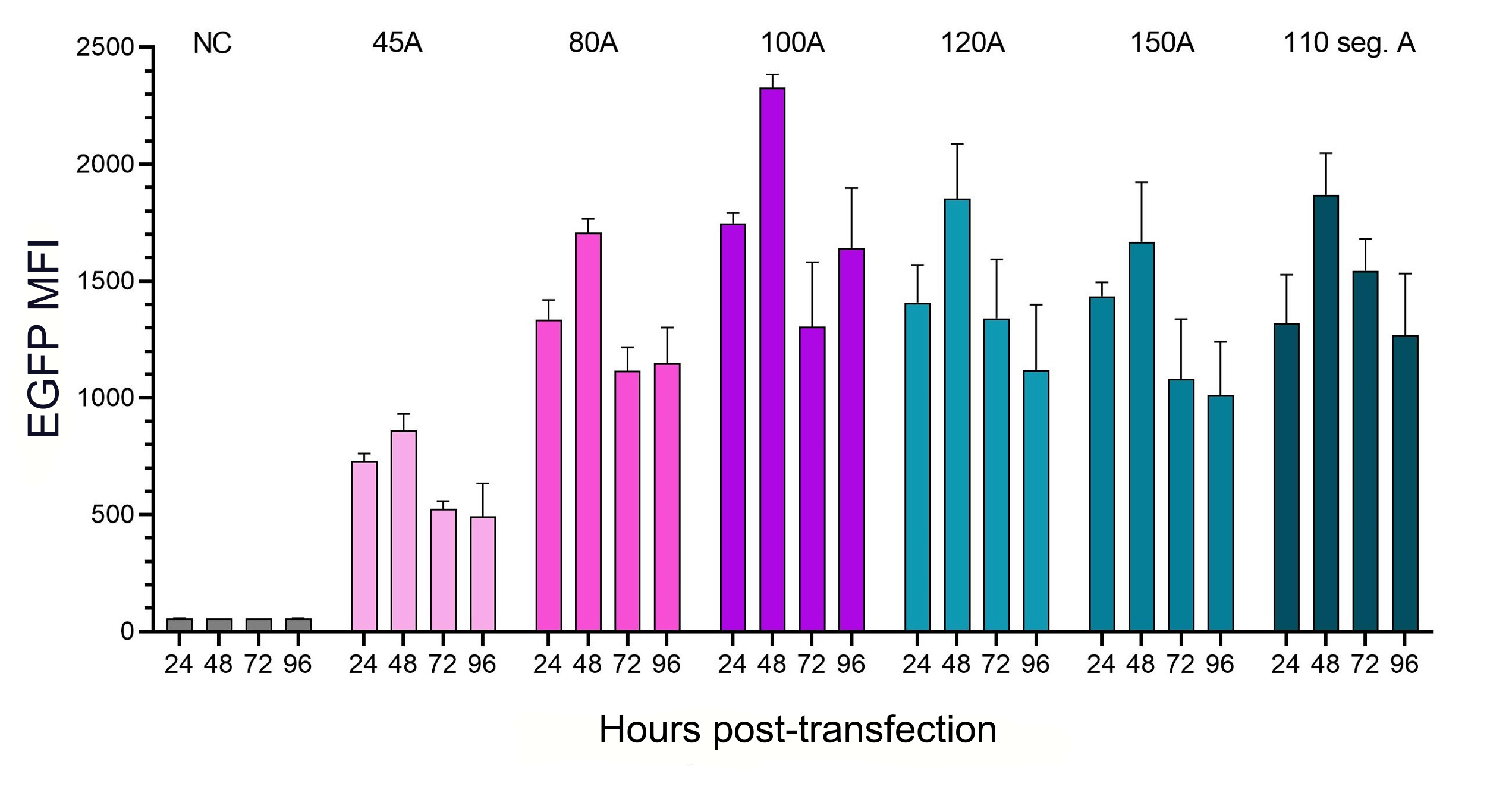
Figure 4. The influence of polyA length and structure on translational efficiency. HEK293T cells were transfected with IVT EGFP mRNA with the same Cap1 and UTRs but different polyA tail lengths as listed above.
IVT RNA Production
- As fast as 5 weeks from vector cloning to LNP encapsulation
- Synthesis of up to 10,000 nt mRNA and saRNA, and 5,000 nt circRNA from ug to gram scales
- High capping efficiency (up to 99%) by co-transcriptional or enzymatic methods
- Various modified nucleotide options: m1Ψ, m5C, 5moU, etc.
- Proprietary purification process to efficiently remove impurities
- Comprehensive QC panel
- We offer cell-free production of IVT template DNA that greatly shortens the production time for large-scale IVT RNA manufacturing.
- mRNA integrity
- Capping efficiency
- Modified nucleotide
- dsRNA removal
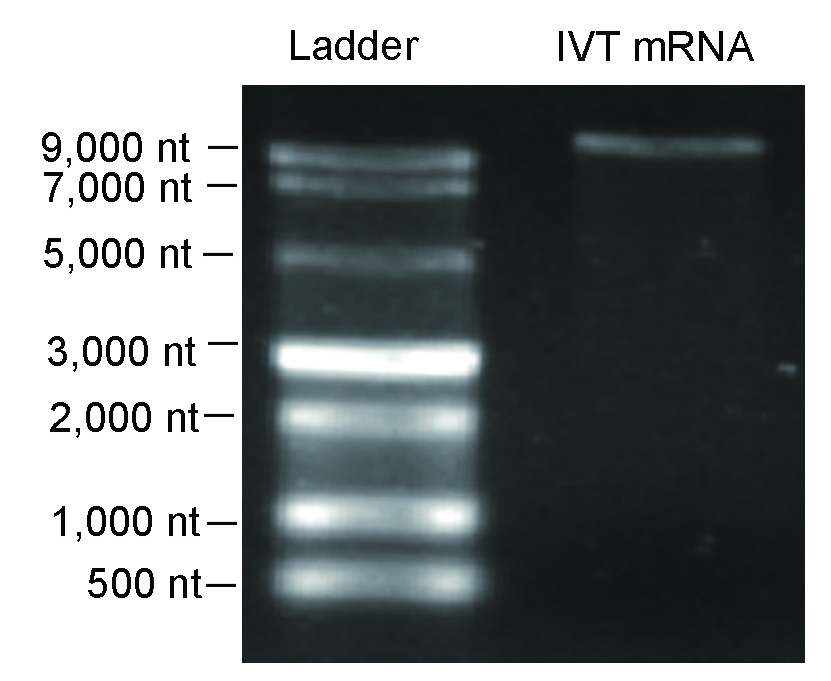
Figure 5. Denaturing agarose gel result indicating high integrity IVT RNA >10,000 nt.
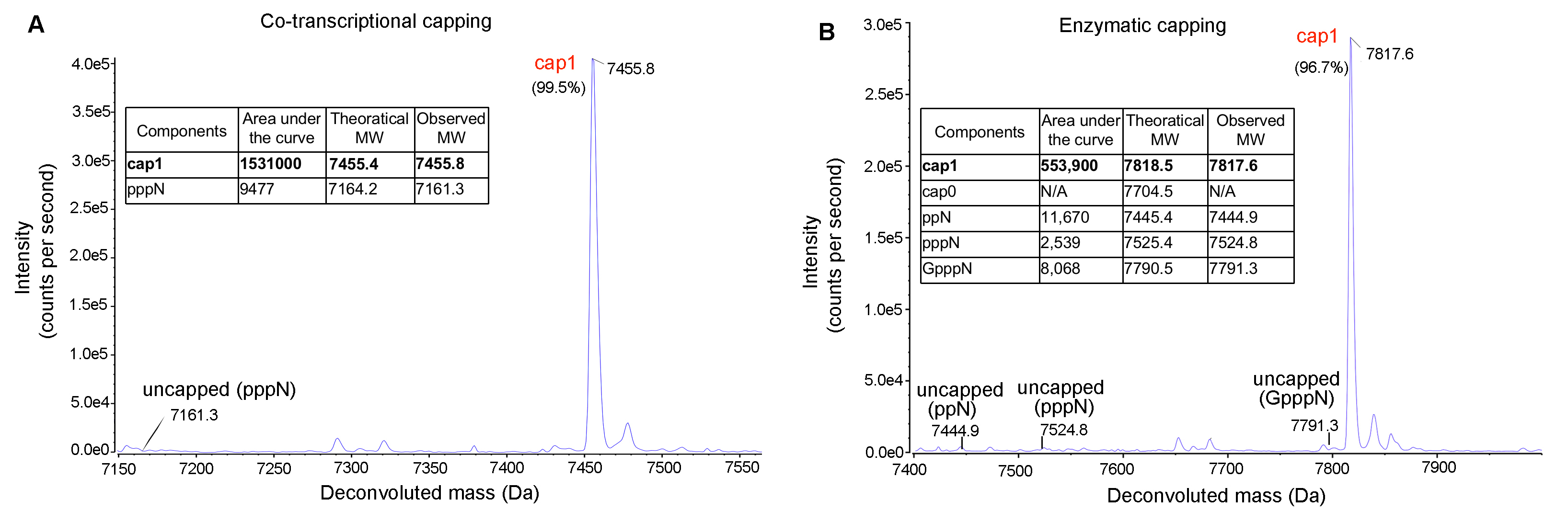
Figure 6. IVT mRNA capping efficiency analyzed by LC-MS. Highly efficient capping (>99%) can be achieved either using (A) co-transcriptional or (B) enzymatic approaches.
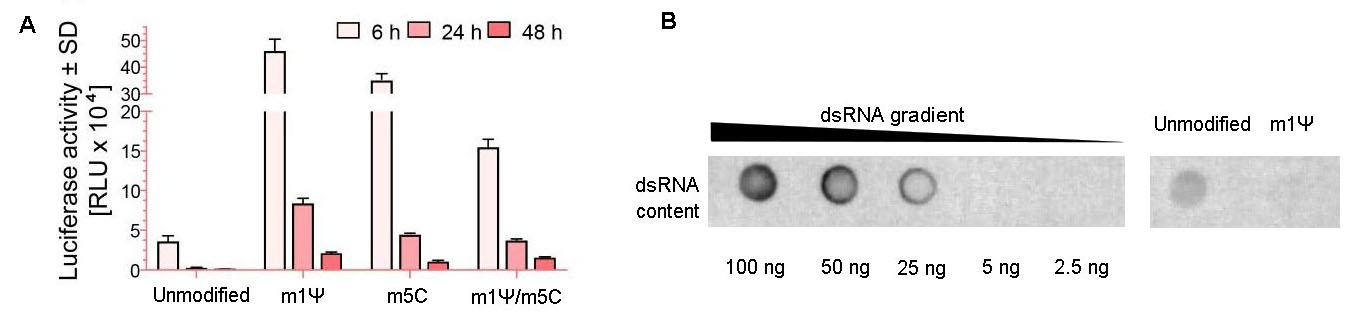
Figure 7. Modified nucleotides increase mRNA expression and decrease dsRNA impurities. (A) Expression of firefly luciferase in HEK293T cells. mRNA was generated with or without modified nucleotides, N1-Methylpseudouridine (m1Ψ) and 5-Methylcytosine (m5C). Cells were grown on 12-well plates and transfected with 1 ug of mRNA per well. Luciferase activities in HEK293T cells at 6 h, 24 h, and 48 h post-transfection were measured. Error bars indicate standard deviations. (B) Equal amounts (750 ng per dot) of magnetic bead-purified EGFP IVT mRNA with or without nucleotide modification (m1Ψ) was assessed by dot blot assay for dsRNA impurities.
Double-stranded RNA (dsRNA) is a by-product of IVT and a major trigger of undesired immunogenicity. The dot blot result demonstrates that extra purification steps (e.g. IP-PR) may be necessary to achieve an ultra-purification scale with very low levels of dsRNA.
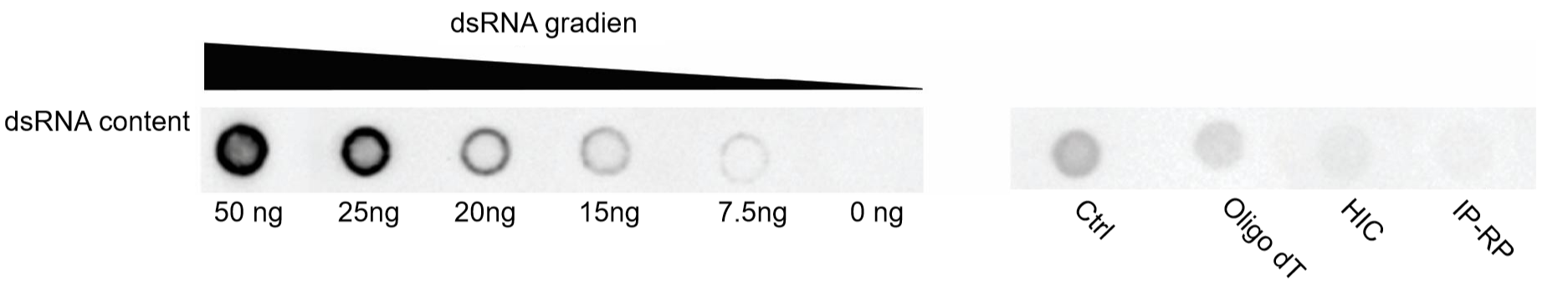
Figure 8. dsRNA removal efficiency of different purification processes. Equal amounts (1500 ng per dot) of hSpCas9 IVT mRNA purified by different processes was assessed by dot blot assay to estimate dsRNA impurities. Abbreviations: HIC, Hydrophobic interaction chromatography; IP-RP, Ion-pair reversed-phase liquid chromatography.
LNP Encapsulation
Visit our LNP Encapsulation service page for more details.
Quality Control (QC)
VectorBuilder offers a full spectrum of QC assays for IVT RNA. Default QC items (marked with √) are always performed while optional QC items are performed depending on individual project needs.
| Attribute | QC assay | Research-grade | GMP-like | |
|---|---|---|---|---|
| Identity | mRNA sequence | Sanger sequencing | √ | √ |
| mRNA length | Denaturing agarose gel electrophoresis | √ | √ | |
| Capillary gel electrophoresis (CGE) | Optional | √ | ||
| General/physical property | mRNA concentration | UV spectrophotometry | √ | √ |
| RiboGreen assay | Optional | √ | ||
| Appearance | Visual inspection | Optional | √ | |
| Potency | Gene expression | In vitro translation followed by Western blot | Optional | Optional |
| Cell transfection | Optional | Optional | ||
| Safety | Sterility | Bioburden test | Optional | √ |
| Mycoplasma | Culture method | Optional | √ | |
| qPCR | Optional | Optional | ||
| Endotoxin | Kinetic chromogenic assay (KCA) | Optional | √ | |
| Purity | mRNA integrity | Denaturing agarose gel electrophoresis | √ | √ |
| Capillary gel electrophoresis (CGE) | Optional | √ | ||
| A260/280 | UV spectrophotometry | √ | √ | |
| Capping efficiency | LC-MS | Optional | √ | |
| PolyA analysis | LC-MS | Optional | √ | |
| Residual dsRNA | Dot blot assay | Optional | √ | |
| Residual plasmid DNA | qPCR | Optional | √ | |
| Residual protein | NanoOrange assay | Optional | √ | |
| Residual solvents | Gas chromatography | Optional | Optional | |
| Circularization efficiency (for circRNA) | Denaturing agarose gel electrophoresis | √ | Optional | |
| Capillary gel electrophoresis (CGE) | Optional | √ | ||
Functional Validation
Our scientists and technical team can help you with functional study to optimize your IVT RNA design for therapeutic purpose, including:
- Assessing the effects of sequence optimization of various vector components (UTR, coding sequence, polyA, Kozak, etc.) in vitro and in vivo
- Testing IVT RNA for various applications, such as antigen presentation, antibody expression, CAR expression, and CRISPR
- Assessing LNP gene delivery efficacy and safety using animal models including rodents and non-human primates (NHPs)
Documents
Brochures & Flyers User InstructionsCertificate of Analysis (COA)
Material Safety Data Sheet (MSDS)