Shine Bright, Little Cell: A Comprehensive Guide to Choosing the Right Reporter Gene
Keywords: Reporter genes, fluorescent reporters, luciferase assay
Overview
Reporters in molecular biology are molecules that are easy to detect and measure. Hence, they are widely used as indicators to study the expression of a gene of interest or the activity of a regulatory region, which can otherwise be challenging to measure directly. Of all the different types of reporters, fluorescent proteins (FP), luciferase, and β-galactosidase have emerged as some of the most popular tools in biological research. The choice of which specific reporter to use depends greatly on your research question — that is, if your data will be quantitative or qualitative and if you are working with live cells or mounted tissues. This review will take a deep dive into each of the three reporters to provide you with a roadmap to select the most appropriate reporter for your specific needs.
To get the reporter genes into your cells of interest, popular gene delivery tools like plasmid transfection, recombinant virus, or mRNA are typically used. (Figure 1).
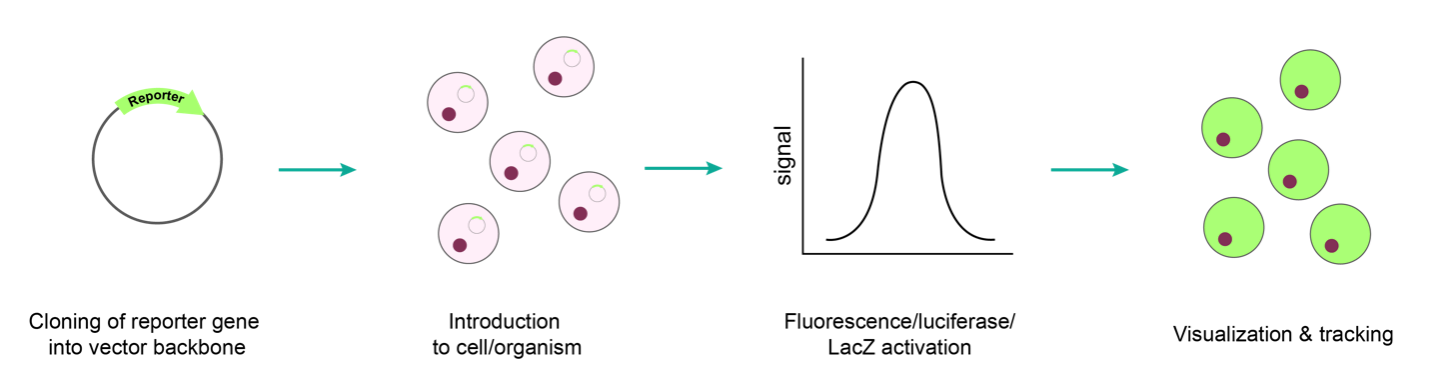
Figure 1. Typical workflow for expression of a reporter gene
Apart from the choice of the right reporter, location of the reporter gene on the vector is crucial and largely depends on the research question being asked. Some examples include:
1. How can I find out if my promoter is active in a certain tissue or cell? A reporter gene can be cloned into a suitable vector backbone downstream of tissue/cell specific promoter sequence.
2. How does the level of expression for my GOI change after a specific treatment? And what should I do if it's hard to measure the expression of my GOI? A reporter gene can be cloned into a suitable vector backbone linked to a target GOI using linkers to create a fusion protein (Read for more information on linkers).
3. How do I measure the strength of my promoter? A quantifiable reporter gene can be cloned into a suitable vector backbone downstream of a promoter of interest allowing for the assessment of promoter activity based on reporter gene expression levels.
If you are unsure about the design of your vector with reporter genes specific to your research question, the VectorBuilder team can help.
Fluorescent protein reporters
The discovery and isolation of green fluorescent protein (GFP) from jellyfish and its use as a transgene in a variety of organisms have been crucial to the progress of molecular biology. GFP’s ability to fold into a functional fluorescent protein in a variety of eukaryotic cell types as well as its ability to be observed easily with conventional fluorescence microscopes have been the reasons for its use in thousands of experiments studying patterns of gene expression, subcellular localization, and other cellular processes.
Since the discovery of GFP, several other FPs have been identified and used in biological research. Some popular examples include Red Fluorescent Protein (RFP) which produces red light; Yellow Fluorescent Protein (YFP), a GFP derivative with a yellow-shifted emission; mCherry known for its intense bright red color; and tagBFP2, a member of the Blue Fluorescent Protein (BFP) family- further information on all of these (and many more FPs!) is available in our fluorescent protein guide.
Fluorescent proteins are ideal as a reporter system, as they provide a view into the cellular and molecular mechanisms of living cells and organisms in real time. Additionally, FPs have a wide range of colors that allow for the simultaneous visualization of multiple cellular components making it easy to study multiple targets, while LacZ and Luciferase are typically limited to a single signal. Fluorescent proteins are, however, prone to photobleaching (the loss of fluorescence with exposure to light) and autofluorescence, thus limiting their use in some cases. This VectorBuilder article provides detailed information about optimizing fluorescent protein markers based on brightness, maturation speed, photostability, and spectral properties for various applications, including single-color, multicolor, and fluorescence resonance energy transfer (FRET) experiments. Additionally, we have optimized and validated an EGFP mRNA for use in zebrafish reporter experiments through sequence engineering (Figure 2).
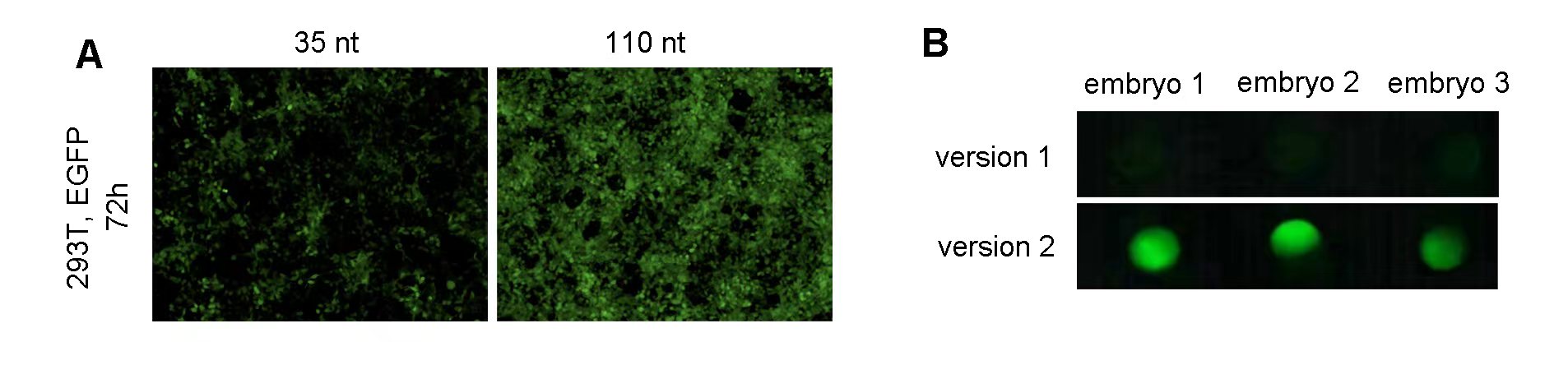
Figure 2. (A) 293T cells grown on a 12-well plate were transfected with 1 ug of zebrafish EGFP mRNA per well. Representative images at 72 h post-transfection indicate that 110 nt polyA tail increases EGFP expression in vitro. (B) One-cell stage zebrafish embryos were screened for EGFP expression at 6 h post-microinjection of 250 pg of zebrafish EGFP IVT mRNA. Fluorescent images indicate that optimized Kozak sequence (version 2) increased EGFP expression in vivo.
Luciferase reporters
Scientific research into bioluminescence began in the early 1900s with experiments on luminous organisms, which led to the discovery that bioluminescence is a chemical reaction involving an enzyme (luciferase) and substrate (luciferin), leading to emission of light that is easily measured. Luciferase assays are a very common use of this reporter, which provide a readout of transgene expression. The bioluminescence in a luciferase assay is a direct read out of activity of the luciferase enzyme; luciferase acts on the specific substrate luciferin in the presence of ATP and oxygen. The light emitted from the reaction of oxidized luciferin can be measured and quantified using a luminometer. The intensity of the light is directly proportional to the amount of luciferase enzyme present, which can be used as an accurate readout to quantify the level of gene expression.
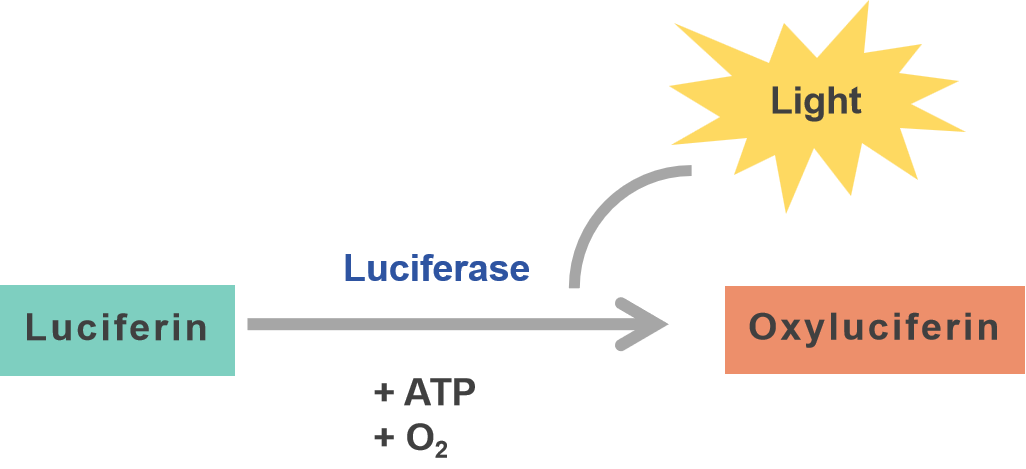
A significant advantage of luciferase is the minimal background signal, which provides highly sensitive quantitative measurements. However, the assay requires the addition of substrates to the reaction, and the transient nature of this signal might eliminate its use in long-term tracking. Luciferase is also an excellent target for optimization, including codon optimization. We have performed sequence optimization of the HiExpressTM IVT firefly luciferase mRNA to achieve high-level expression (Figure 3).
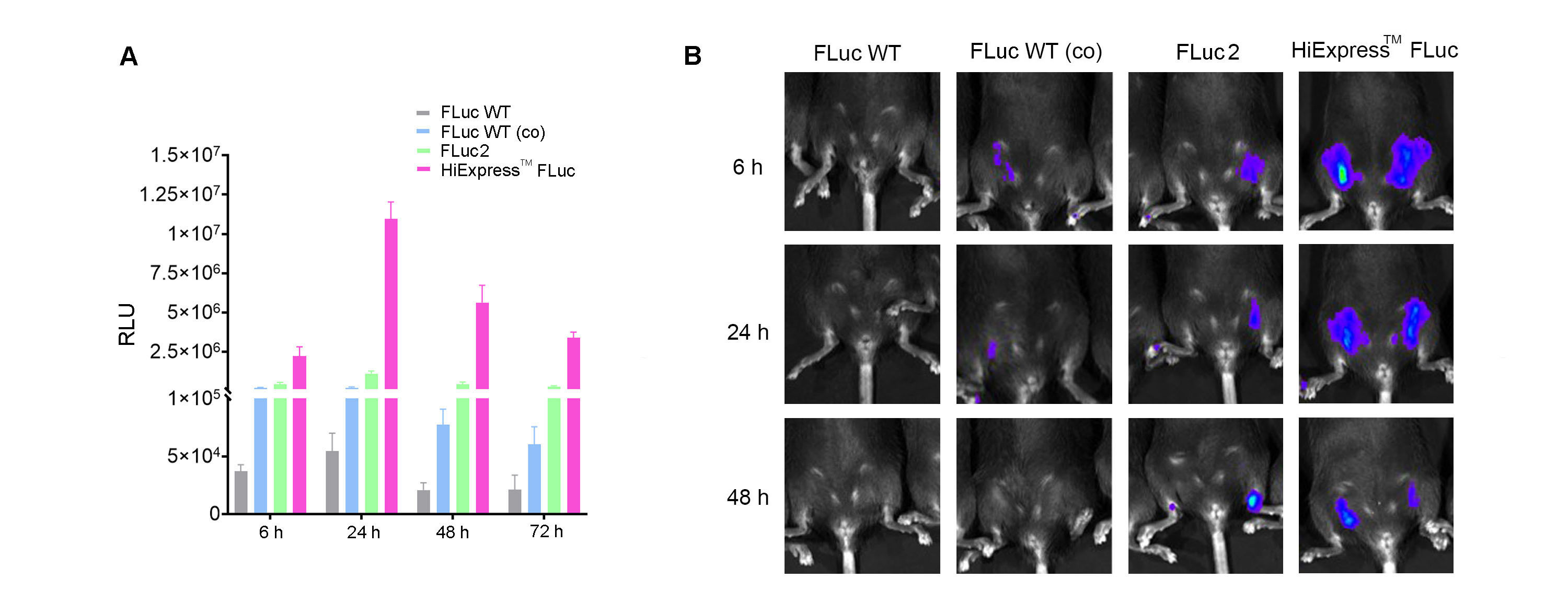
Figure 3. (A) Expression of HiExpressTM Firefly Luciferase mRNA and other luciferase mRNA in HEK293T cells. Cells grown on a 12-well plate were transfected with 0.5 ug of mRNA per well and luciferase activity was measured at 6 h, 24 h, 48 h, and 72 h post transfection. (B) Luciferase activity measured in adult C57BL/6 mice injected intramuscularly with 30 ug of LNP encapsulated mRNA at 6 h, 24 h, and 48 h post injection. FLuc WT indicates wild-type firefly luciferase. FLuc WT (co) indicates codon-optimized wild-type firefly luciferase. FLuc2 indicates Luc2 firefly luciferase.
β-galactosidase/ LacZ reporter system
The lacZ gene, which encodes the enzyme β-galactosidase (β-gal) necessary for lactose metabolism in E. coli, was a pivotal enzyme in early studies on transcriptional regulation. Following in-depth studies of the lacZ gene, it achieved widespread adoption as a reporter gene in biological research. β-gal is a tetrameric enzyme that normally breaks down lactose; however, when it acts on X-gal, a derivate of lactose, the byproducts of the reaction result in an easily identifiable blue color and therefore can be used to detect enzymatic activity. The expression of β-gal can be easily monitored by adding X-gal to the host cell, where the blue appearance of the cells is used as a readout to identify gene expression. The LacZ system is predominantly used for the identification of positive clones of recombinant bacteria in cloning experiments and to confirm promoter activity in various organisms. Additionally, by fusing constructs of the GOI with the lacZ gene it is possible to study protein localization, protein-protein interactions, and the effects of components like Cre-responsive FLEX elements on gene expression.
β-gal/Lac-Z is a cost-effective system, providing enzymatic activity that gives a high signal-to-noise ratio and allows the detection of low-level gene expression. It has a very broad host-range, including organisms from bacteria to mammalian cells. The enzyme is non-toxic which allows for gene expression studies in cells and whole organisms. However, conducting β-gal assays in live cells and organisms is complicated and many experiments require cellular lysis or fixation of cells/organisms. Additionally, this assay is less quantitative compared to fluorescent proteins or luciferase assays.
Summary
| Reporter System | Type of Reaction | Ideal Use Cases | Advantages | Limitations |
|---|---|---|---|---|
| Fluorescent Proteins | Non-enzymatic |
|
|
|
| Luciferase | Enzymatic |
|
|
|
| β-Galactosidase (LacZ) | Enzymatic |
|
|
|
It should be noted that the different reporters can be used in conjunction in experiments, leveraging the unique advantages of each reporter system to provide comprehensive biological insights. For example, utilizing these reporters in different cellular compartments (e.g., nucleus for one reporter and cytoplasm for another) can allow for more precise localization studies. The simultaneous use of these reporters, however, requires careful experimental design to avoid cross-interference and ensure accurate interpretation of the results.
Keywords: Reporter genes, Fluorescent proteins, Luciferase, β-galactosidase, GFP, RFP, YFP, mCherry, BFP, Cloning and expression of reporter genes, Bioluminescent imaging tools, Promoter activity assessment, Vector design for gene expression, In vivo and in vitro reporter gene assays, Quantitative and qualitative analysis.
References
1. Kremers, Gert-Jan, Sarah G. Gilbert, Paula J. Cranfill, Michael W. Davidson, and David W. Piston. 2011. "Fluorescent Proteins at a Glance." *Journal of Cell Science* 124 (2): 157–160.
2. Chudakov, Dmitriy M., Mikhail V. Matz, Sergey Lukyanov, and Konstantin A. Lukyanov. 2010. "Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues." *Physiological Reviews* 90 (3): 1103–1163.
3. Cranfill, P., B. Sell, M. Baird, et al. 2016. "Quantitative Assessment of Fluorescent Proteins." *Nature Methods* 13: 557–562.
4. Yeh, H.W. and H.W. Ai. 2019. "Development and Applications of Bioluminescent and Chemiluminescent Reporters and Biosensors." *Annual Review of Analytical Chemistry* (Palo Alto, Calif) 12 (1): 129–150. [https://doi.org/10.1146/annurev-anchem-061318-115027](https://doi.org/10.1146/annurev-anchem-061318-115027). Epub 2019 Feb 20. PMID: 30786216; PMCID: PMC6565457.
5. Zambito, Giorgia, Chintan Chawda, and Laura Mezzanotte. 2021. "Emerging Tools for Bioluminescence Imaging." *Current Opinion in Chemical Biology* 63: 86–94. ISSN 1367-5931. [https://doi.org/10.1016/j.cbpa.2021.02.005].
6. Sharma, Shiv K., Sijan Poudel Sharma, and Roger M. Leblanc. 2021. "Methods of Detection of β-galactosidase Enzyme in Living Cells." *Enzyme and Microbial Technology* 150: 109885. ISSN 0141-0229. [https://doi.org/10.1016/j.enzmictec.2021.109885].
7. Sedzro, DM, SMF Bellah, H Akbar, and SMS Billah. 2018. "Structure, Function, Application and Modification Strategy of β –Galactosidase." *Journal of Multidisciplinary Research Review* 1 (1): 10–16.



